Magnesium »
PDB 8uee-8un4 »
8uis »
Magnesium in PDB 8uis: Structure of Transcription Complex Pol II-Dsif-Nelf-Tfiis
Enzymatic activity of Structure of Transcription Complex Pol II-Dsif-Nelf-Tfiis
All present enzymatic activity of Structure of Transcription Complex Pol II-Dsif-Nelf-Tfiis:
2.7.7.6;
2.7.7.6;
Other elements in 8uis:
The structure of Structure of Transcription Complex Pol II-Dsif-Nelf-Tfiis also contains other interesting chemical elements:
| Zinc | (Zn) | 8 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of Transcription Complex Pol II-Dsif-Nelf-Tfiis
(pdb code 8uis). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of Transcription Complex Pol II-Dsif-Nelf-Tfiis, PDB code: 8uis:
In total only one binding site of Magnesium was determined in the Structure of Transcription Complex Pol II-Dsif-Nelf-Tfiis, PDB code: 8uis:
Magnesium binding site 1 out of 1 in 8uis
Go back to
Magnesium binding site 1 out
of 1 in the Structure of Transcription Complex Pol II-Dsif-Nelf-Tfiis
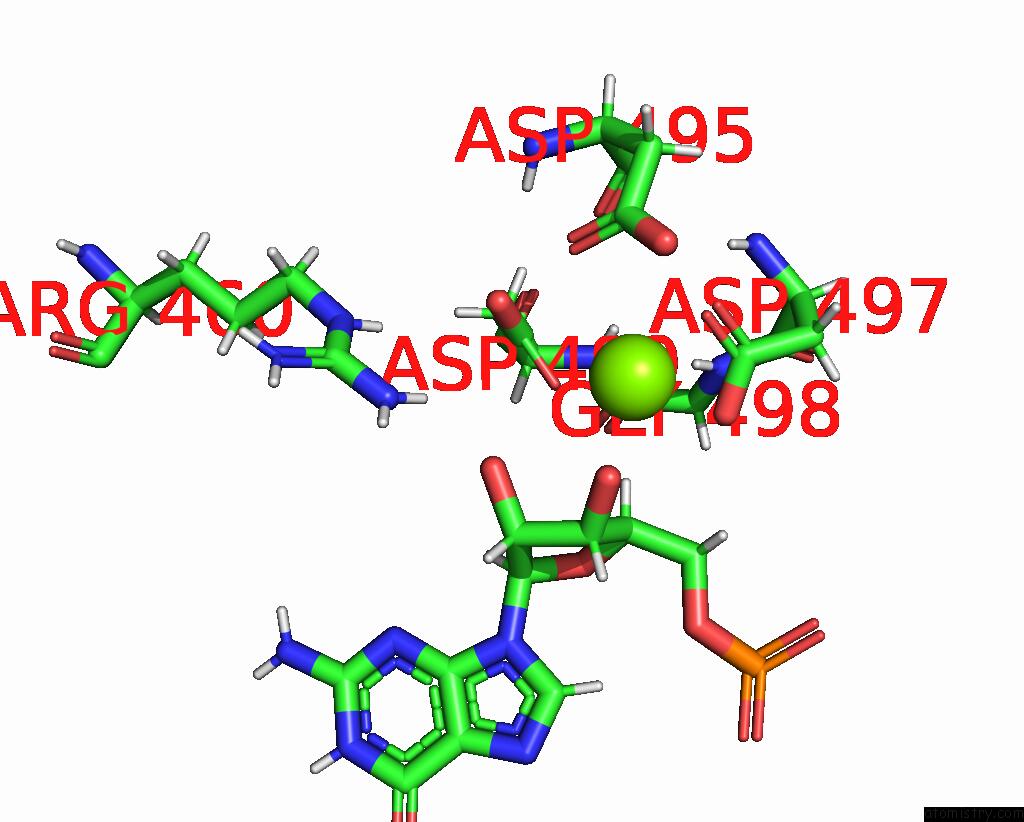
Mono view
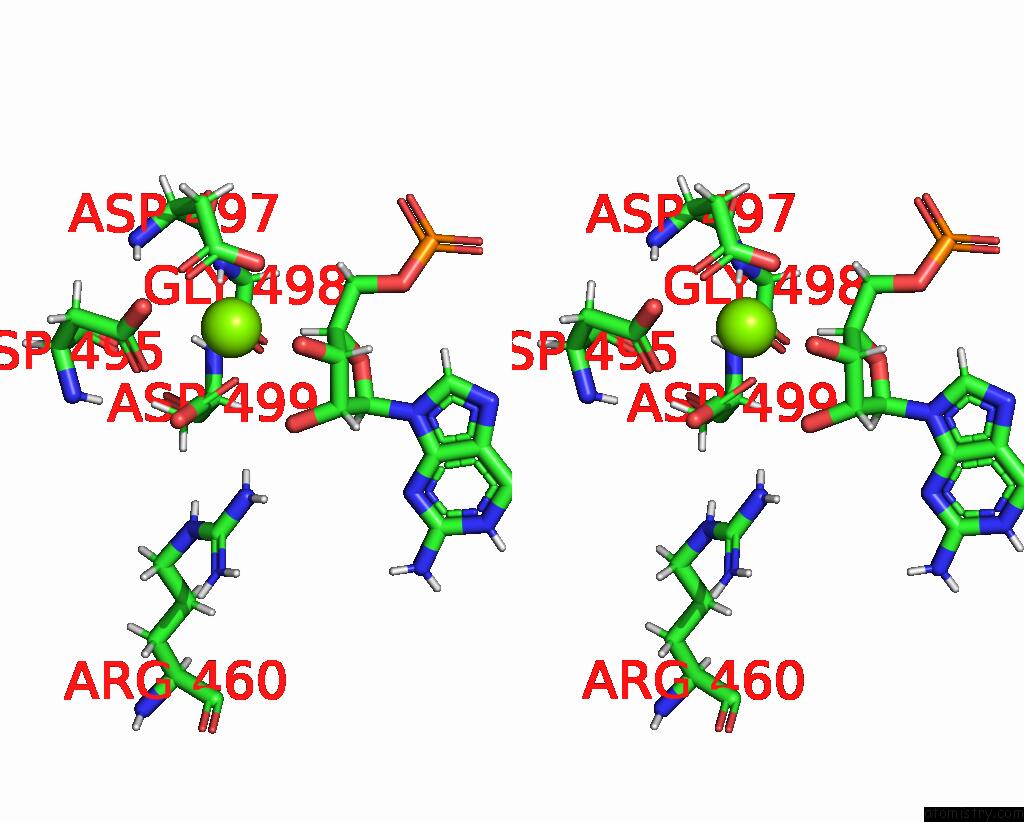
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Transcription Complex Pol II-Dsif-Nelf-Tfiis within 5.0Å range:
|
Reference:
B.G.Su,
S.M.Vos.
Distinct Negative Elongation Factor Conformations Regulate Rna Polymerase II Promoter-Proximal Pausing. Mol.Cell 2024.
ISSN: ISSN 1097-2765
PubMed: 38401543
DOI: 10.1016/J.MOLCEL.2024.01.023
Page generated: Fri Aug 15 16:57:06 2025
ISSN: ISSN 1097-2765
PubMed: 38401543
DOI: 10.1016/J.MOLCEL.2024.01.023
Last articles
Br in 9R0QBr in 9J73
Br in 9BJ5
Br in 8Y72
Au in 9D33
As in 9O9I
Al in 9GSG
Zr in 1XC1
Zr in 6Y7P
Zr in 6GNL