Magnesium »
PDB 8y9n-8ynr »
8yih »
Magnesium in PDB 8yih: Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State
Enzymatic activity of Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State
All present enzymatic activity of Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State:
3.1.21.1; 3.1.26.3; 3.6.1.3;
3.1.21.1; 3.1.26.3; 3.6.1.3;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State
(pdb code 8yih). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State, PDB code: 8yih:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State, PDB code: 8yih:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 8yih
Go back to
Magnesium binding site 1 out
of 2 in the Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State
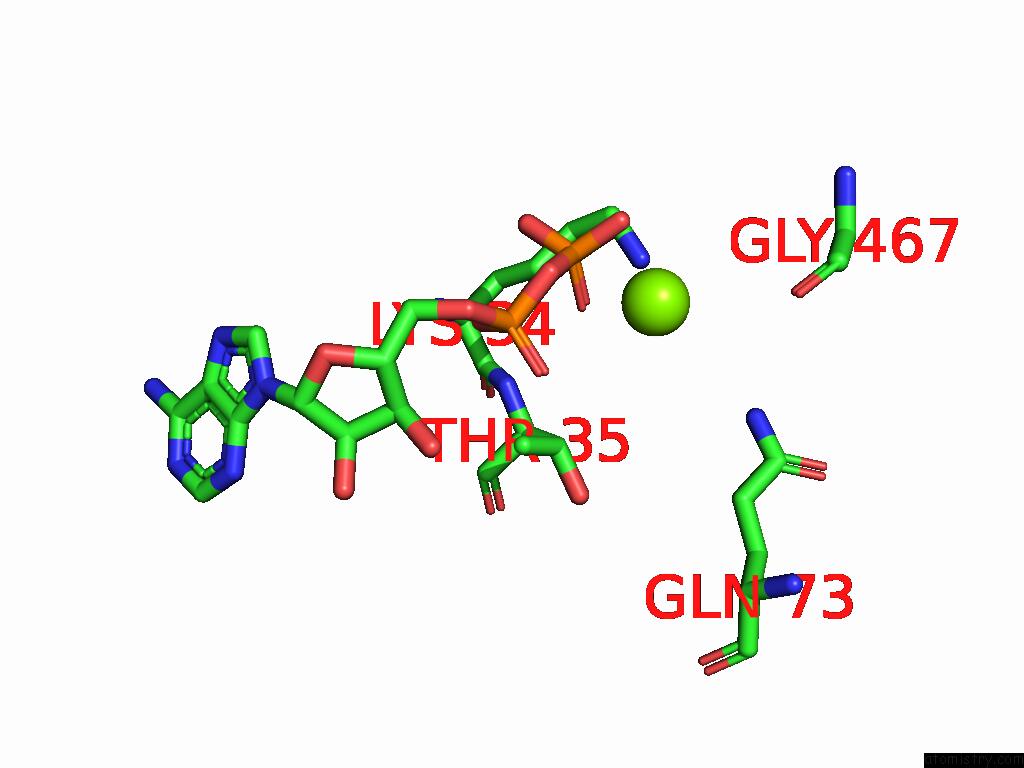
Mono view
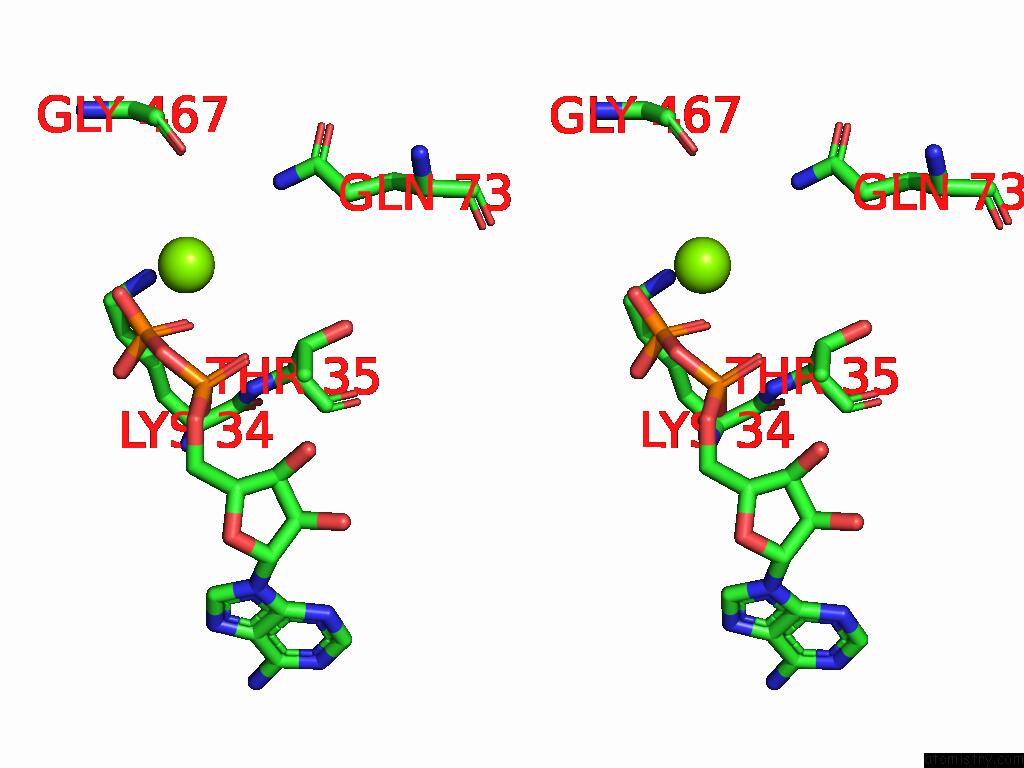
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 8yih
Go back to
Magnesium binding site 2 out
of 2 in the Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State
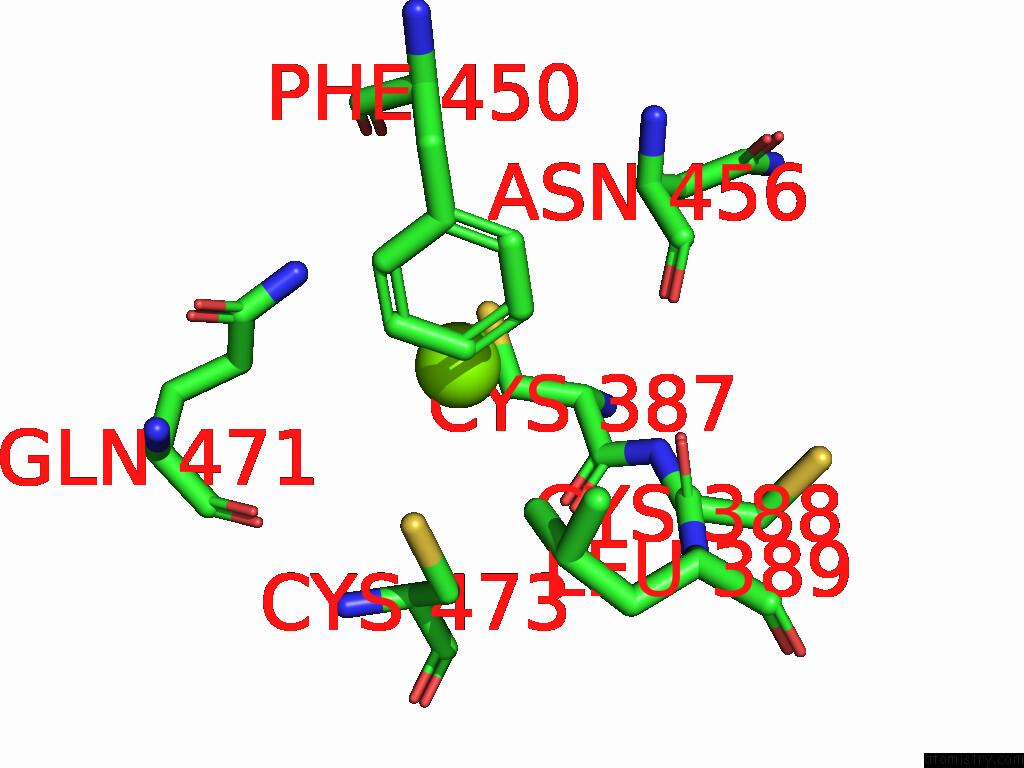
Mono view
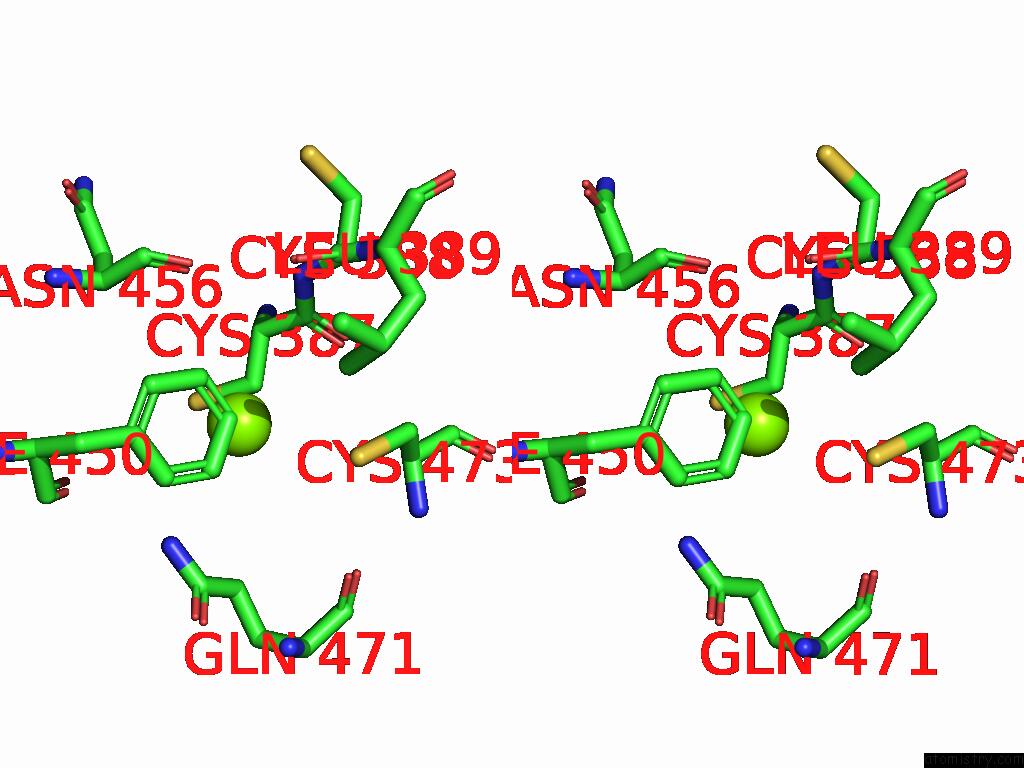
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Dmdcr-2/Loqspd/SLM1 in Pre-Dicing State within 5.0Å range:
|
Reference:
N.Cao,
S.Su,
J.Wang,
J.Ma,
H.-W.Wang.
Structural Basic of Endo-Sirna Processing By Drosophila Dcr-2 and Loqs-Pd To Be Published.
Page generated: Fri Aug 15 21:29:43 2025
Last articles
Mg in 9G09Mg in 9G08
Mg in 9FZF
Mg in 9FZ4
Mg in 9FYX
Mg in 9FYB
Mg in 9FXK
Mg in 9FXN
Mg in 9FX7
Mg in 9FX6