Magnesium »
PDB 9bm1-9bw1 »
9bv1 »
Magnesium in PDB 9bv1: M2A Midnolin-Proteasome (Translocating)
Enzymatic activity of M2A Midnolin-Proteasome (Translocating)
All present enzymatic activity of M2A Midnolin-Proteasome (Translocating):
3.4.25.1;
3.4.25.1;
Other elements in 9bv1:
The structure of M2A Midnolin-Proteasome (Translocating) also contains other interesting chemical elements:
| Zinc | (Zn) | 1 atom |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the M2A Midnolin-Proteasome (Translocating)
(pdb code 9bv1). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the M2A Midnolin-Proteasome (Translocating), PDB code: 9bv1:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the M2A Midnolin-Proteasome (Translocating), PDB code: 9bv1:
Jump to Magnesium binding site number: 1; 2; 3; 4;
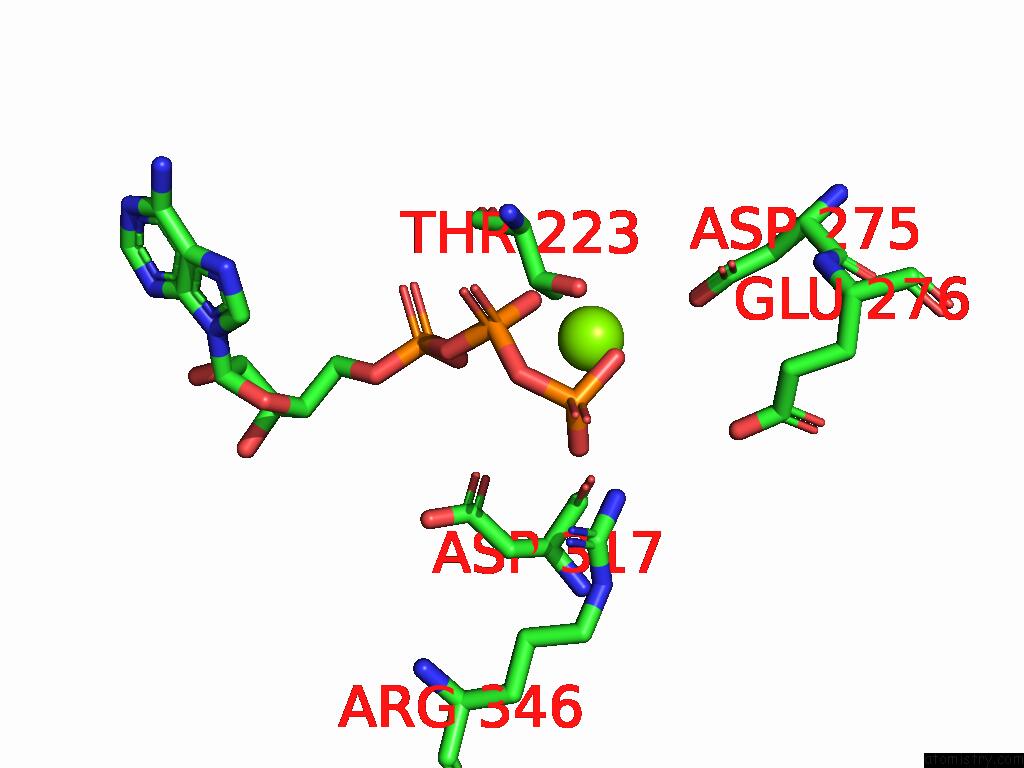
Magnesium binding site 1 out of 4 in 9bv1
Go back to
Magnesium binding site 1 out
of 4 in the M2A Midnolin-Proteasome (Translocating)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of M2A Midnolin-Proteasome (Translocating) within 5.0Å range:
|
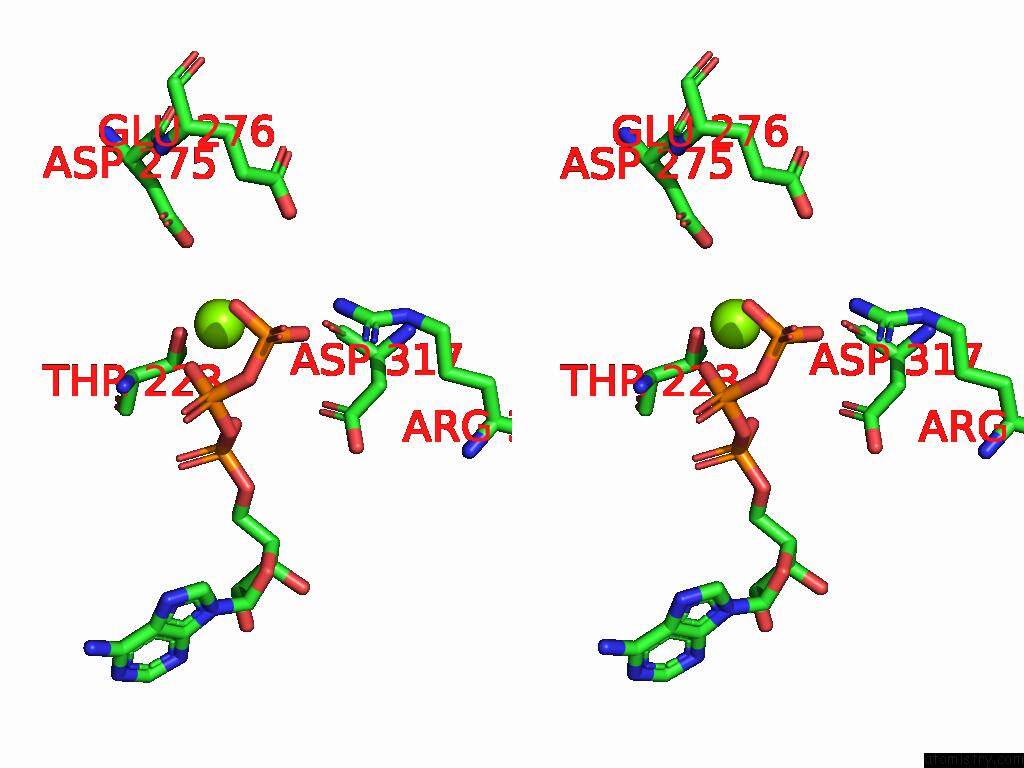
Magnesium binding site 2 out of 4 in 9bv1
Go back to
Magnesium binding site 2 out
of 4 in the M2A Midnolin-Proteasome (Translocating)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of M2A Midnolin-Proteasome (Translocating) within 5.0Å range:
|
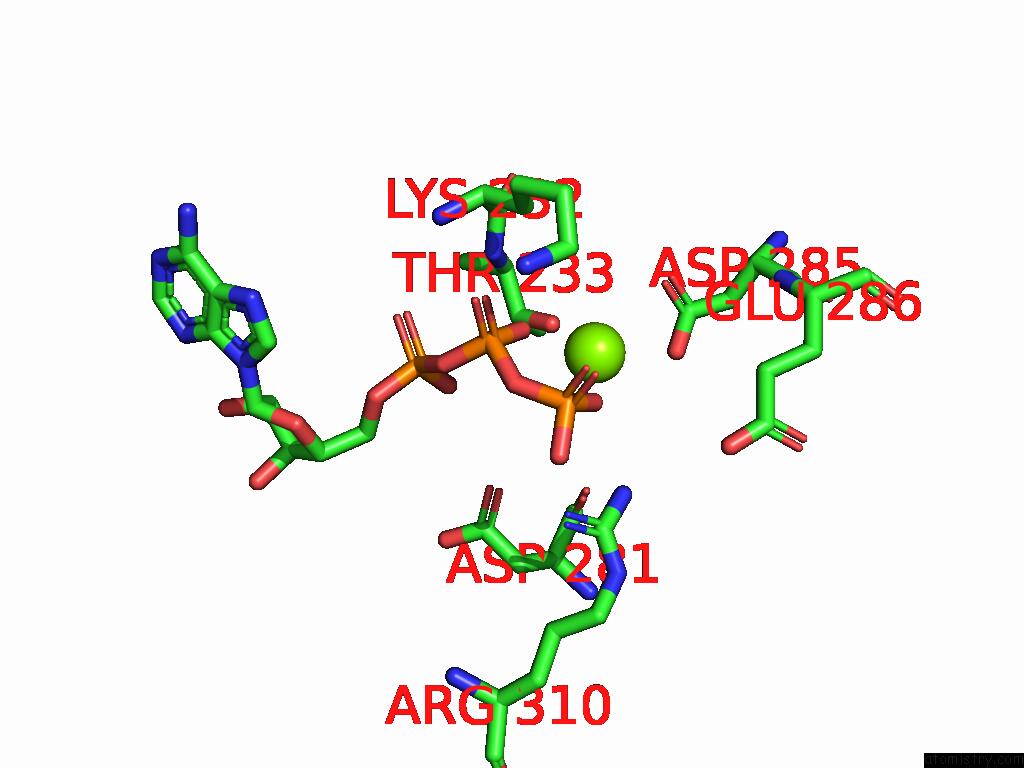
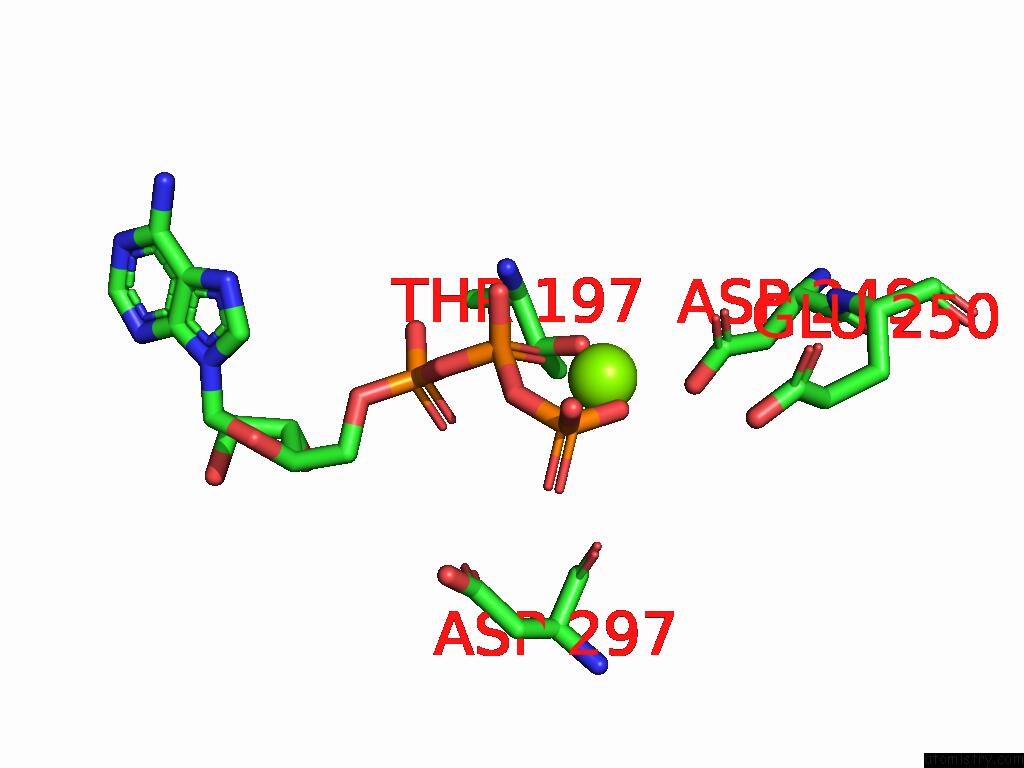
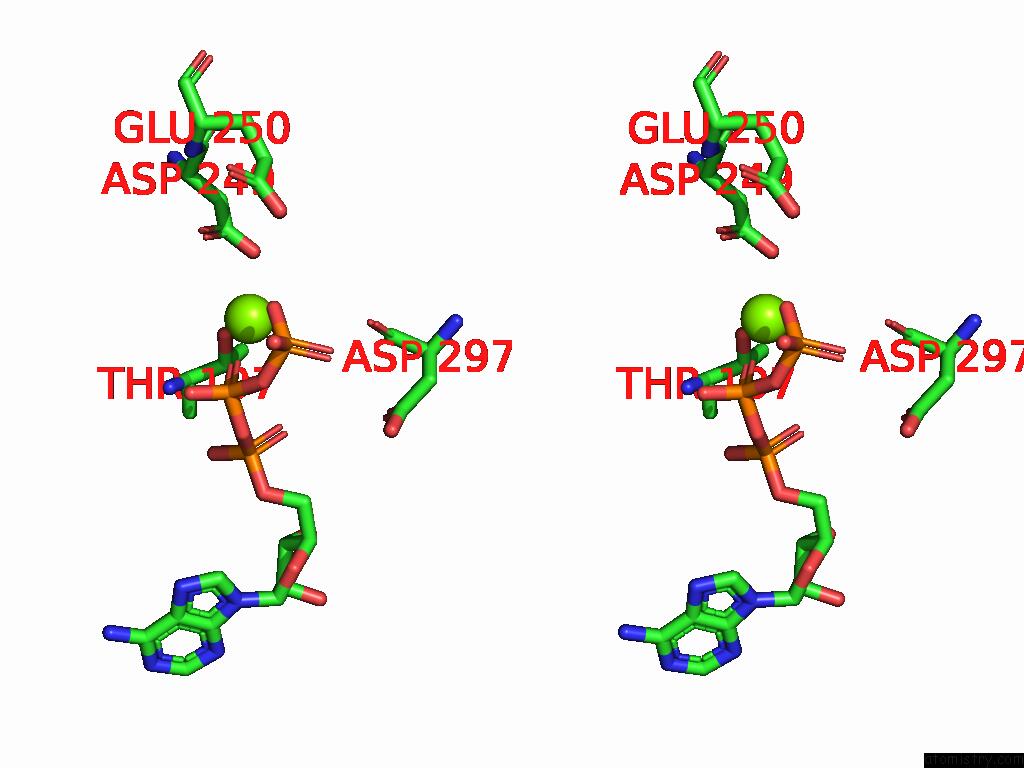
Magnesium binding site 3 out of 4 in 9bv1
Go back to
Magnesium binding site 3 out
of 4 in the M2A Midnolin-Proteasome (Translocating)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of M2A Midnolin-Proteasome (Translocating) within 5.0Å range:
|
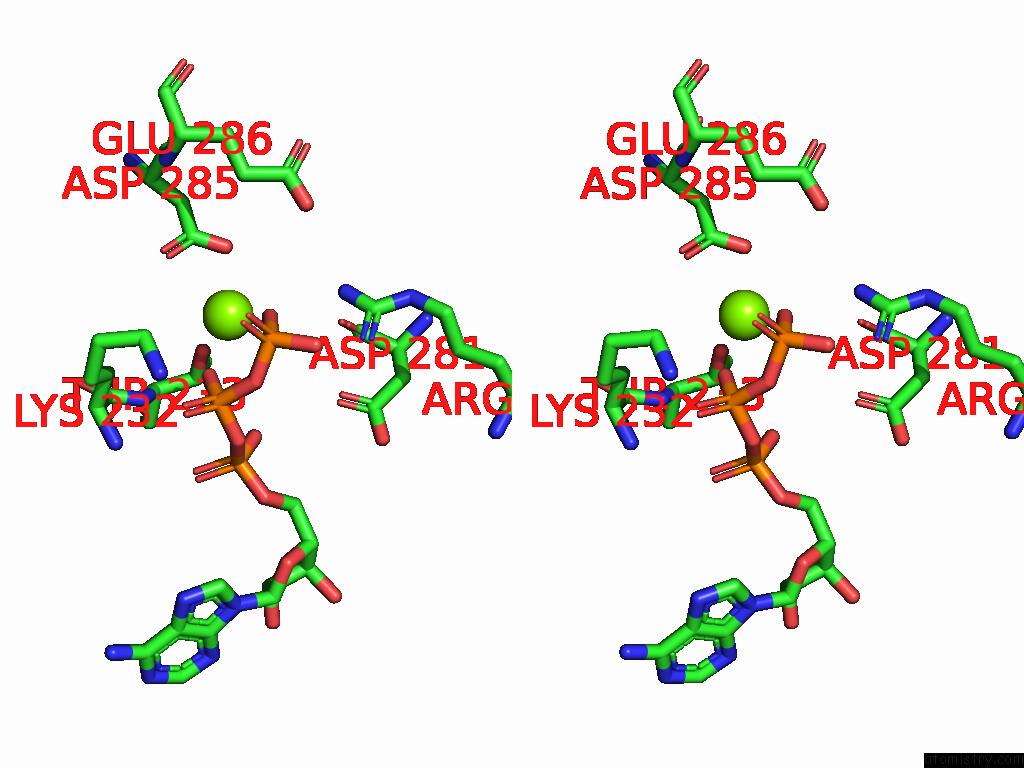
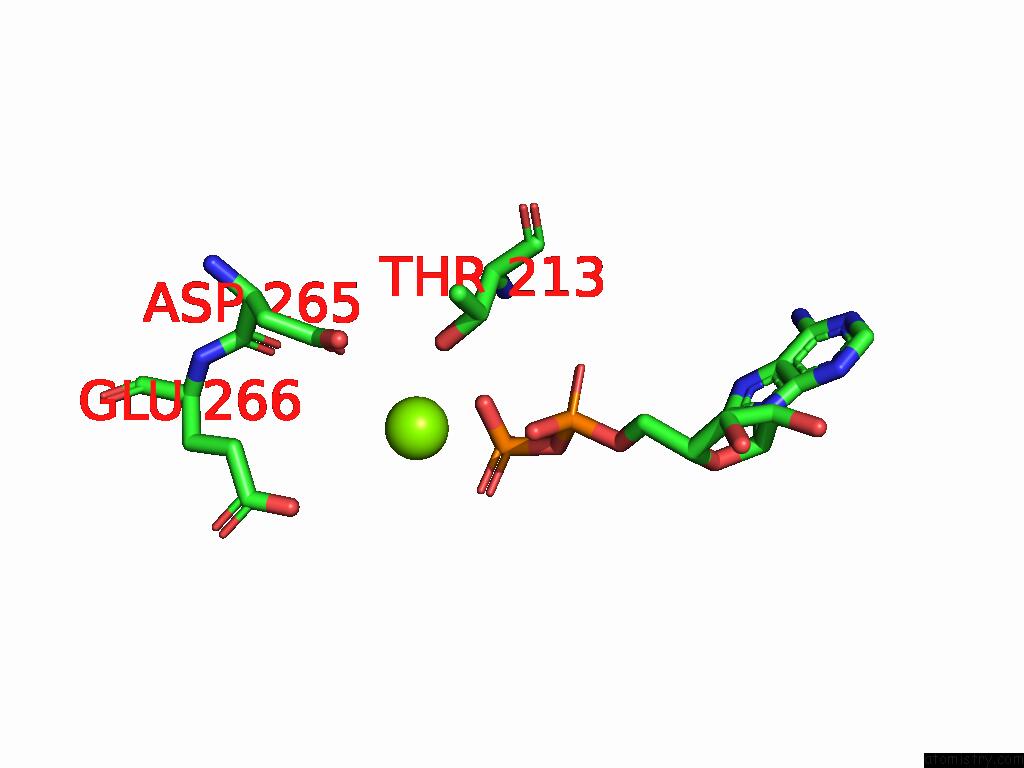
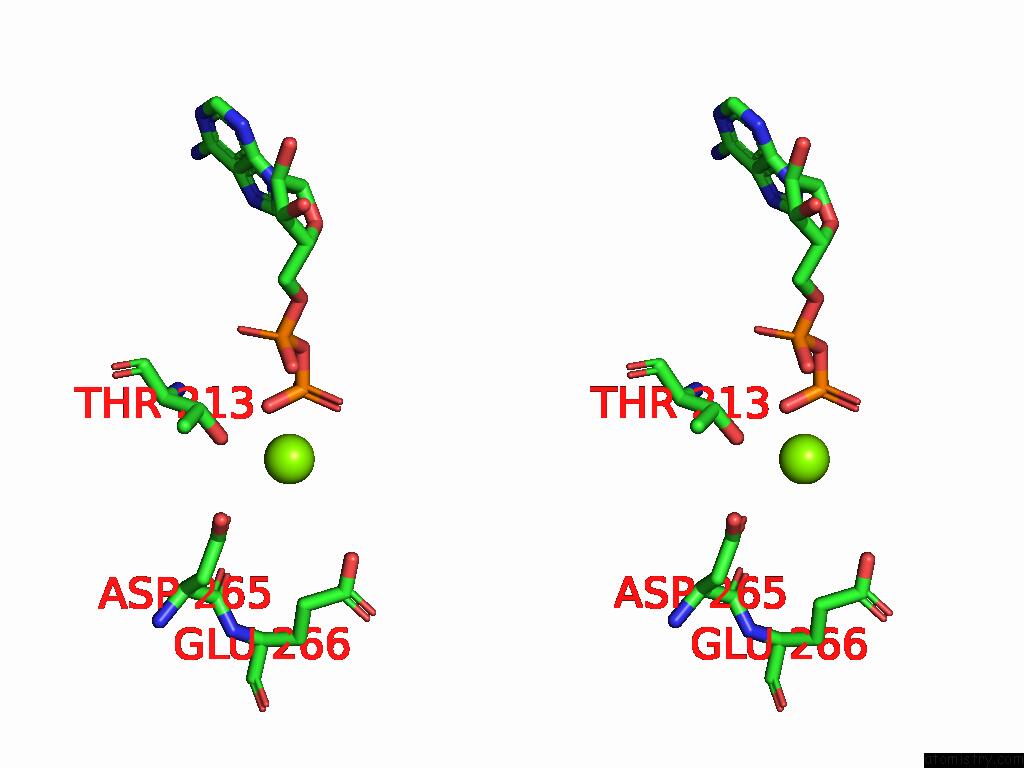
Magnesium binding site 4 out of 4 in 9bv1
Go back to
Magnesium binding site 4 out
of 4 in the M2A Midnolin-Proteasome (Translocating)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of M2A Midnolin-Proteasome (Translocating) within 5.0Å range:
|
Reference:
C.Nardone,
X.Gu,
J.Gao,
M.C.J.Yip,
H.Seo,
K.Song,
L.Sebastian,
S.Dhe-Paganon,
M.Negasi,
N.Kamitaki,
M.E.Greenberg,
S.J.Elledge,
S.Shao.
Structural Basis For Nuclear Protein Degradation By the Midnolin-Proteasome Pathway To Be Published.
Page generated: Fri Aug 15 23:28:48 2025
Last articles
Mn in 5O1UMn in 5NTH
Mn in 5NTG
Mn in 5NSQ
Mn in 5NRZ
Mn in 5NMP
Mn in 5NHA
Mn in 5NPK
Mn in 5NPP
Mn in 5NNB