Magnesium »
PDB 9bz9-9c87 »
9c51 »
Magnesium in PDB 9c51: Cryo-Em Structure of the Strand Displacement Complex (IV) of Yeast Mitochondrial Dna Polymerase Gamma (MIP1) with Downstream Dna
Enzymatic activity of Cryo-Em Structure of the Strand Displacement Complex (IV) of Yeast Mitochondrial Dna Polymerase Gamma (MIP1) with Downstream Dna
All present enzymatic activity of Cryo-Em Structure of the Strand Displacement Complex (IV) of Yeast Mitochondrial Dna Polymerase Gamma (MIP1) with Downstream Dna:
2.7.7.7;
2.7.7.7;
Magnesium Binding Sites:
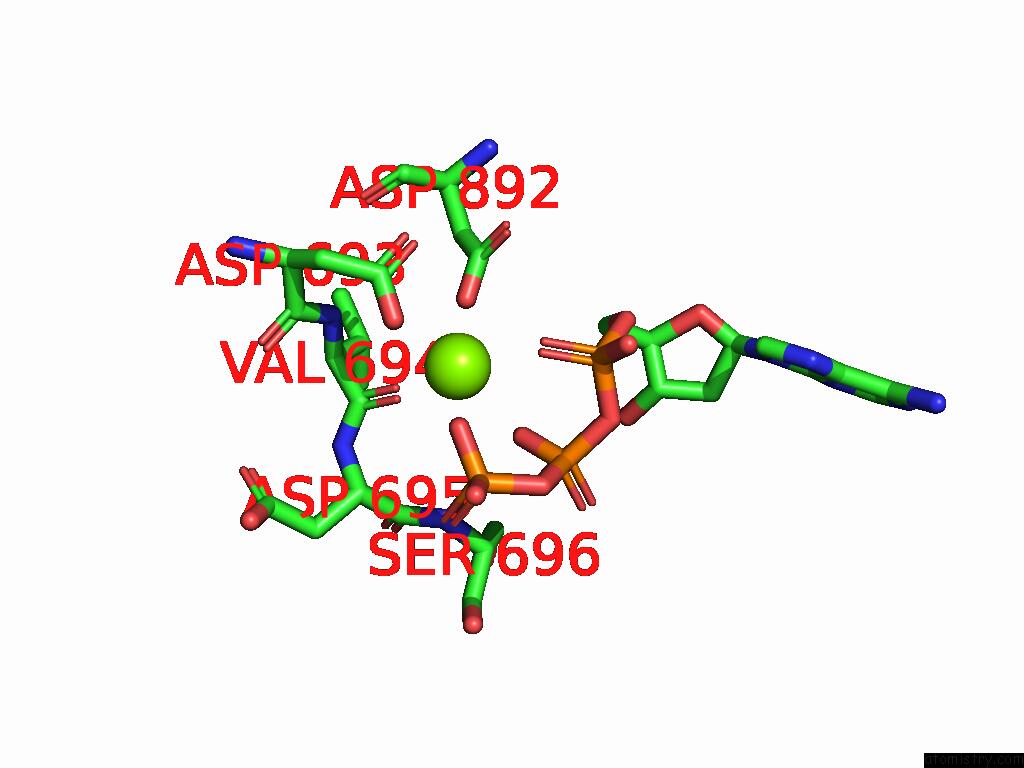
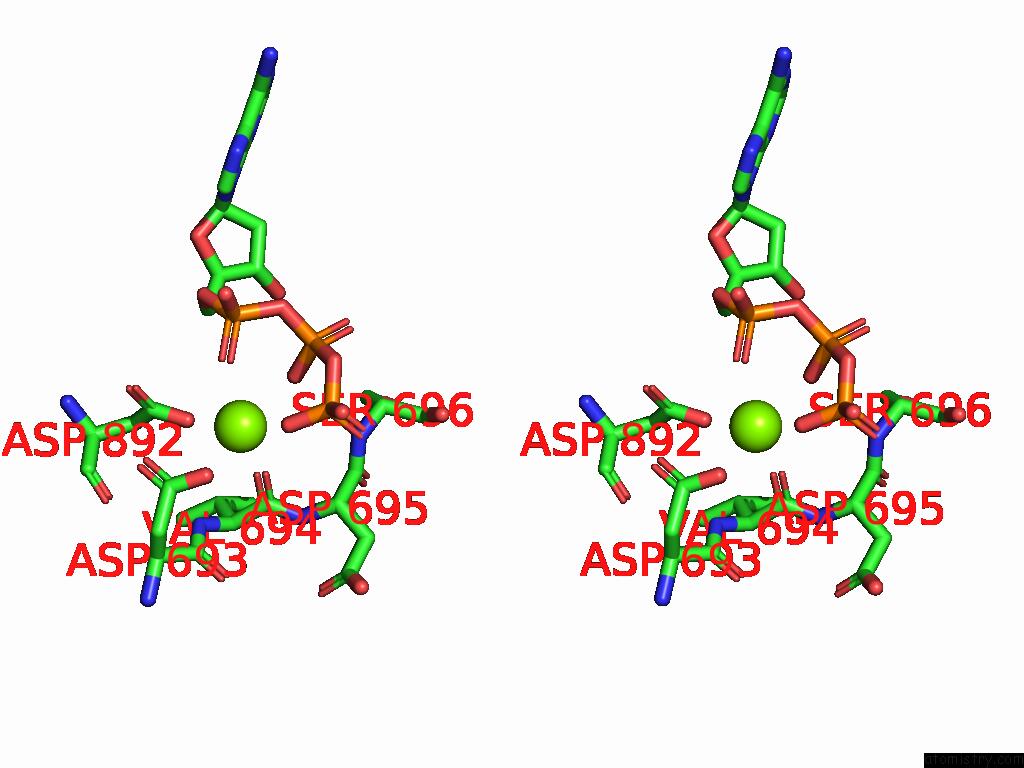
The binding sites of Magnesium atom in the Cryo-Em Structure of the Strand Displacement Complex (IV) of Yeast Mitochondrial Dna Polymerase Gamma (MIP1) with Downstream Dna
(pdb code 9c51). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of the Strand Displacement Complex (IV) of Yeast Mitochondrial Dna Polymerase Gamma (MIP1) with Downstream Dna, PDB code: 9c51:
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of the Strand Displacement Complex (IV) of Yeast Mitochondrial Dna Polymerase Gamma (MIP1) with Downstream Dna, PDB code: 9c51:
Magnesium binding site 1 out of 1 in 9c51
Go back to
Magnesium binding site 1 out
of 1 in the Cryo-Em Structure of the Strand Displacement Complex (IV) of Yeast Mitochondrial Dna Polymerase Gamma (MIP1) with Downstream Dna
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of the Strand Displacement Complex (IV) of Yeast Mitochondrial Dna Polymerase Gamma (MIP1) with Downstream Dna within 5.0Å range:
|
Reference:
A.R.Nayak,
V.Sokolova,
S.Sillamaa,
K.Herbine,
J.Sedman,
D.Temiakov.
Structural Basis For Intrinsic Strand Displacement Activity of Mitochondrial Dna Polymerase Nat Commun V. 16 2417 2025.
ISSN: ESSN 2041-1723
DOI: 10.1038/S41467-025-57594-Z
Page generated: Fri Aug 15 23:37:41 2025
ISSN: ESSN 2041-1723
DOI: 10.1038/S41467-025-57594-Z
Last articles
Mn in 2QGIMn in 2QEY
Mn in 2QF1
Mn in 2QCS
Mn in 2PYO
Mn in 2QEW
Mn in 2QB0
Mn in 2QB5
Mn in 2QB6
Mn in 2Q96