Magnesium »
PDB 9c88-9ck5 »
9cdg »
Magnesium in PDB 9cdg: MORC2 Atpase Dead Mutant - S87A
Other elements in 9cdg:
The structure of MORC2 Atpase Dead Mutant - S87A also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the MORC2 Atpase Dead Mutant - S87A
(pdb code 9cdg). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the MORC2 Atpase Dead Mutant - S87A, PDB code: 9cdg:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the MORC2 Atpase Dead Mutant - S87A, PDB code: 9cdg:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 9cdg
Go back to
Magnesium binding site 1 out
of 3 in the MORC2 Atpase Dead Mutant - S87A
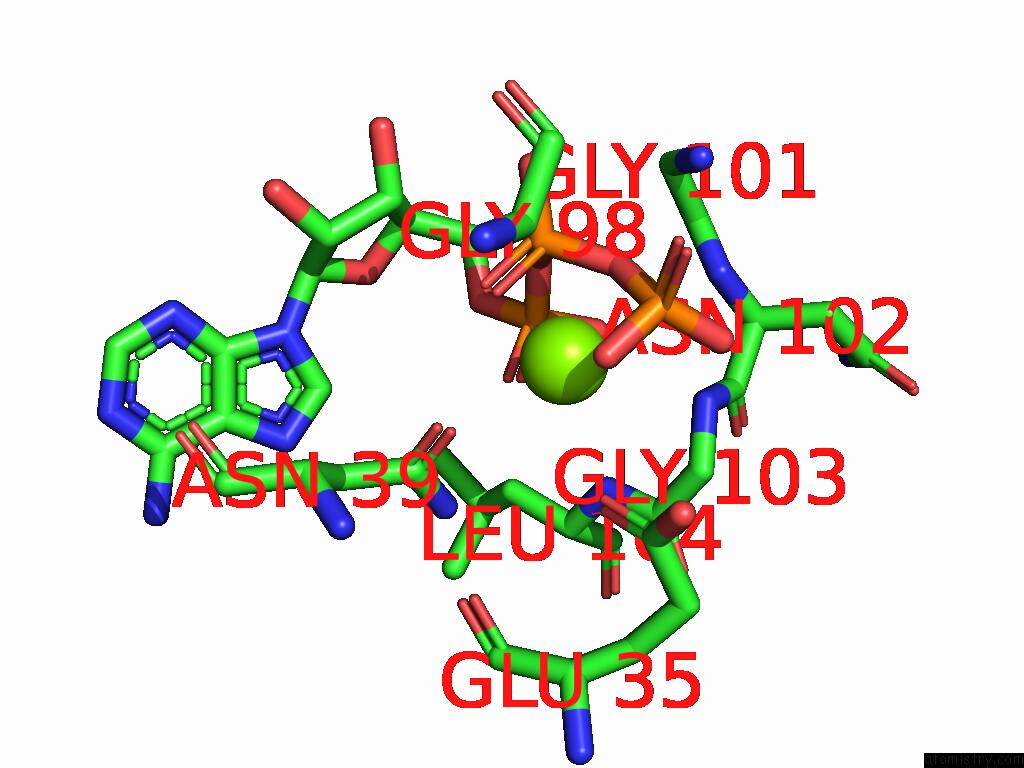
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of MORC2 Atpase Dead Mutant - S87A within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 9cdg
Go back to
Magnesium binding site 2 out
of 3 in the MORC2 Atpase Dead Mutant - S87A
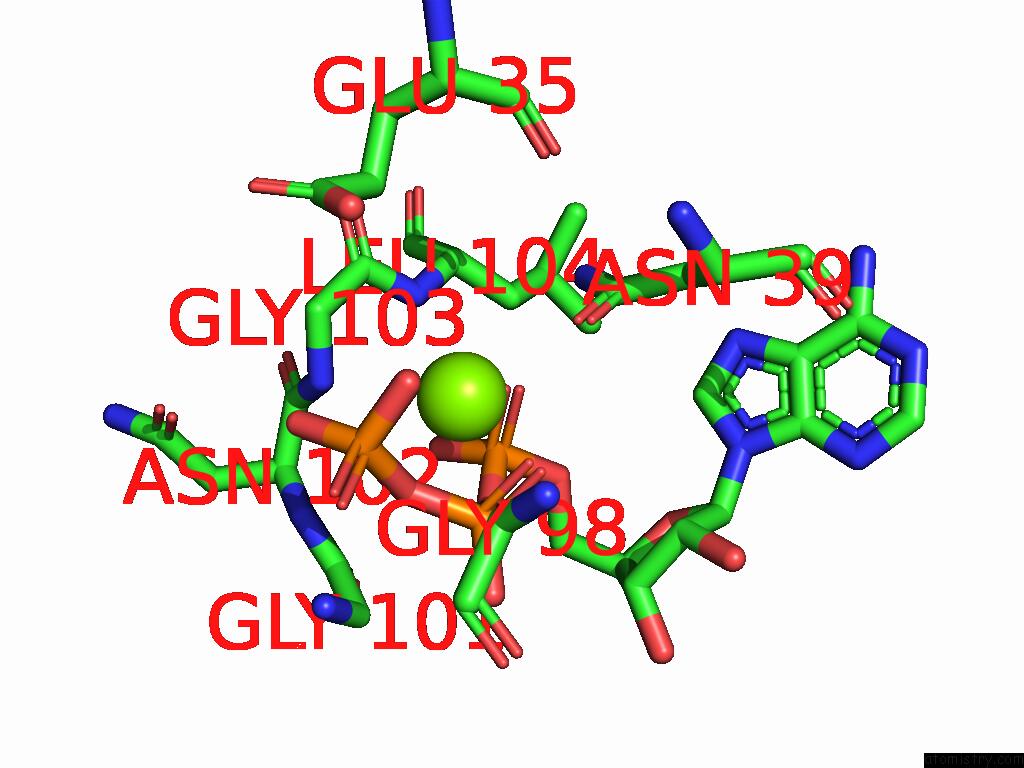
Mono view
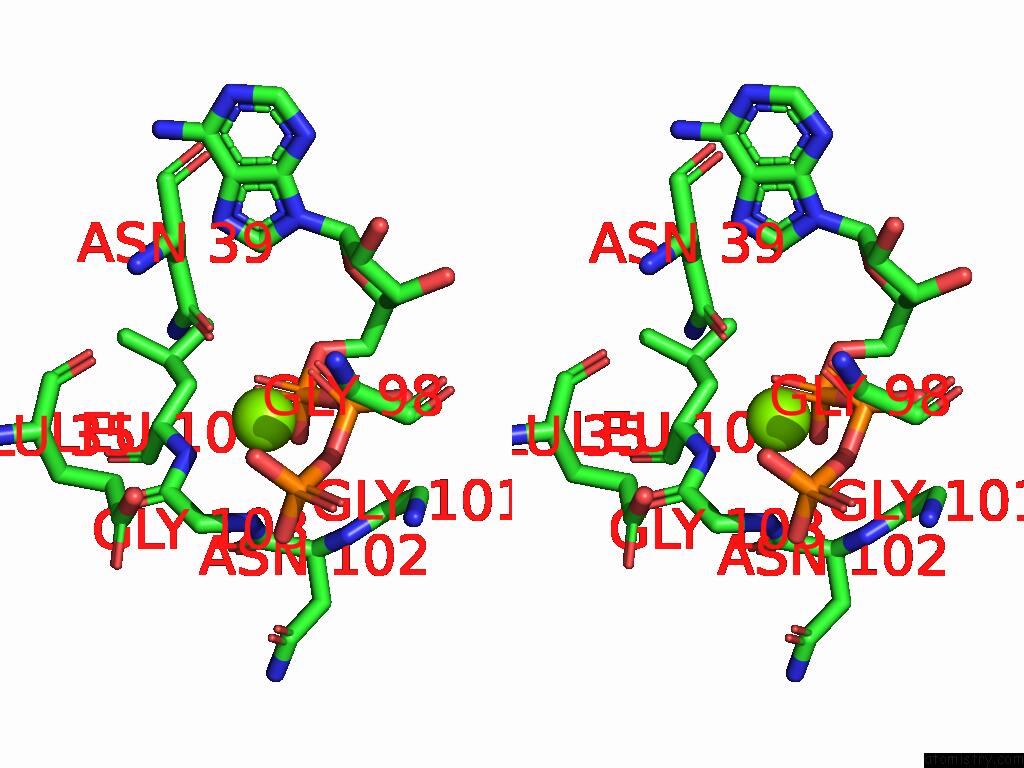
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of MORC2 Atpase Dead Mutant - S87A within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 9cdg
Go back to
Magnesium binding site 3 out
of 3 in the MORC2 Atpase Dead Mutant - S87A
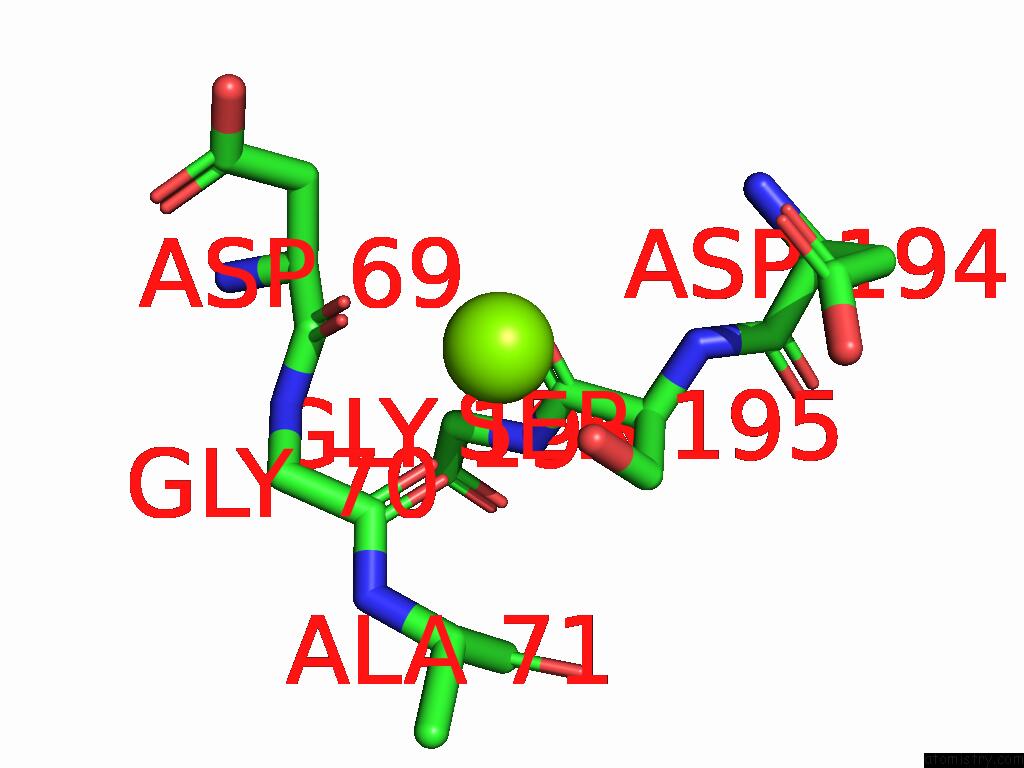
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of MORC2 Atpase Dead Mutant - S87A within 5.0Å range:
|
Reference:
W.Tan,
J.Park,
H.Venugopal,
J.Lou,
P.S.Dias,
P.L.Baldoni,
K.W.Moon,
T.A.Dite,
C.R.Keenan,
A.D.Gurzau,
J.Lee,
T.M.Johanson,
A.Leis,
J.Yousef,
V.Vaibhav,
L.F.Dagley,
C.S.Ang,
L.D.Corso,
C.Davidovich,
S.J.Vervoort,
G.K.Smyth,
M.E.Blewitt,
R.S.Allan,
E.Hinde,
S.D'arcy,
J.K.Ryu,
S.Shakeel.
MORC2 Is A Phosphorylation-Dependent Dna Compaction Machine. Nat Commun V. 16 5606 2025.
ISSN: ESSN 2041-1723
PubMed: 40593625
DOI: 10.1038/S41467-025-60751-Z
Page generated: Sat Aug 16 00:07:11 2025
ISSN: ESSN 2041-1723
PubMed: 40593625
DOI: 10.1038/S41467-025-60751-Z
Last articles
Na in 3RMONa in 3RMN
Na in 3RMM
Na in 3RML
Na in 3RM2
Na in 3RM0
Na in 3RLY
Na in 3RJK
Na in 3RLW
Na in 3RJJ