Magnesium »
PDB 9c88-9ck5 »
9cdv »
Magnesium in PDB 9cdv: Crystal Structure of Rhombotarget A
Protein crystallography data
The structure of Crystal Structure of Rhombotarget A, PDB code: 9cdv
was solved by
A.K.Bera,
S.Rettie,
A.Kang,
G.Bhardwaj,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.71 / 2.00 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 57.156, 106.477, 71.305, 90, 107.04, 90 |
| R / Rfree (%) | 19.5 / 23.4 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Rhombotarget A
(pdb code 9cdv). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Crystal Structure of Rhombotarget A, PDB code: 9cdv:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Crystal Structure of Rhombotarget A, PDB code: 9cdv:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 9cdv
Go back to
Magnesium binding site 1 out
of 3 in the Crystal Structure of Rhombotarget A
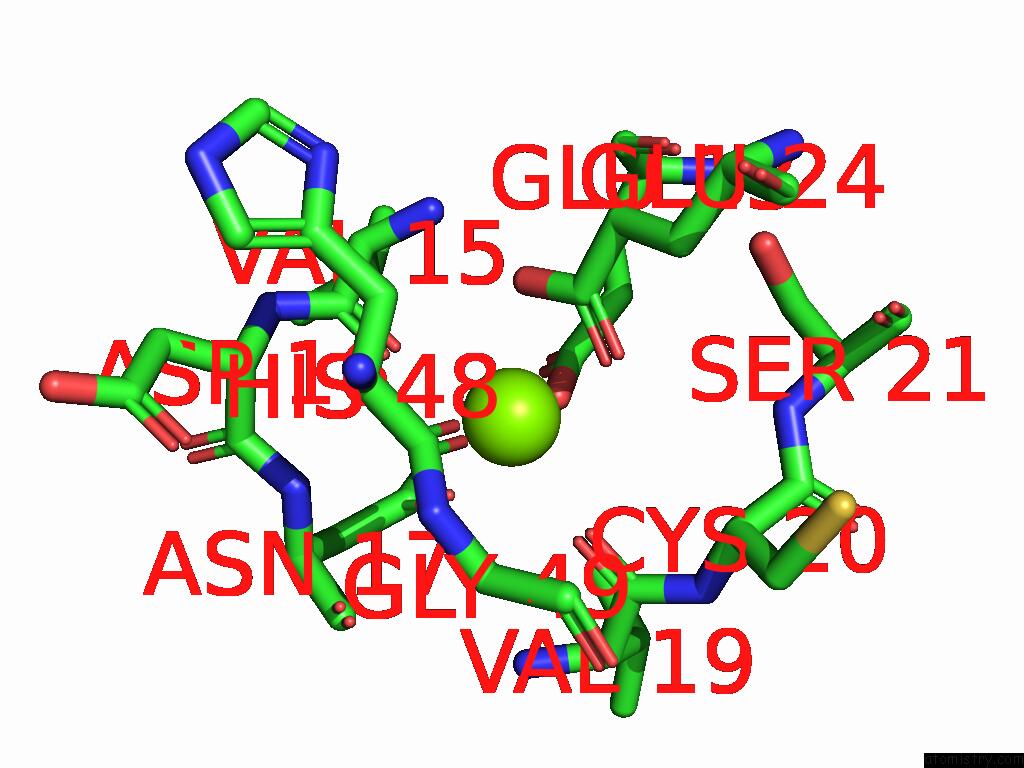
Mono view
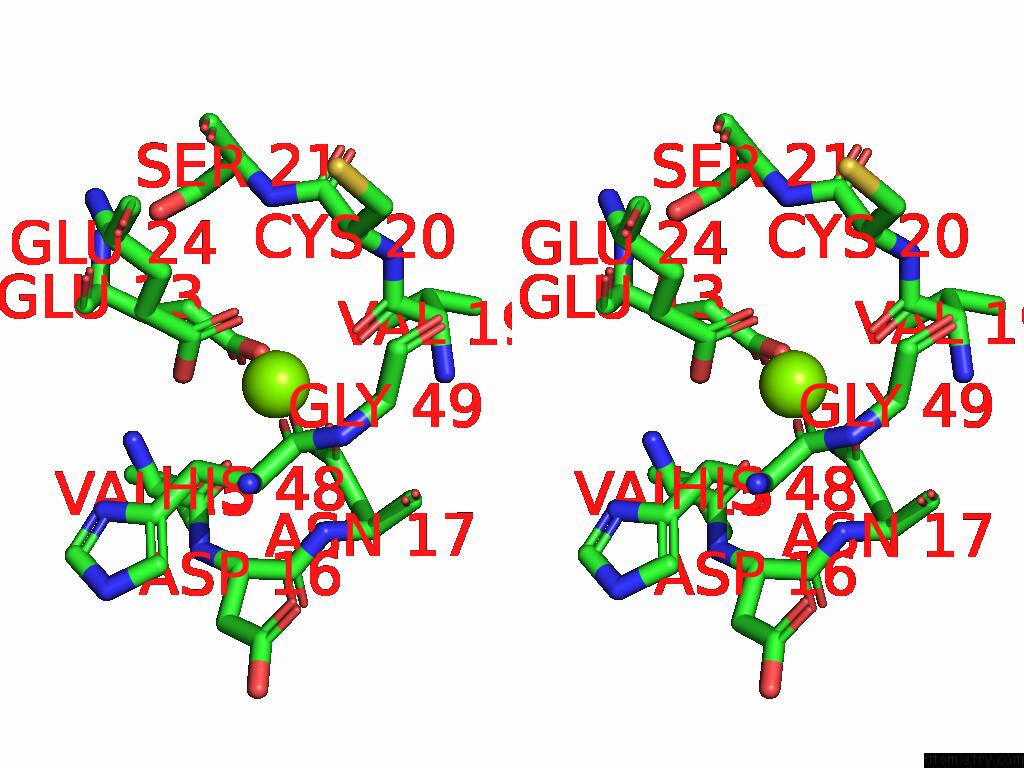
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Rhombotarget A within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 9cdv
Go back to
Magnesium binding site 2 out
of 3 in the Crystal Structure of Rhombotarget A
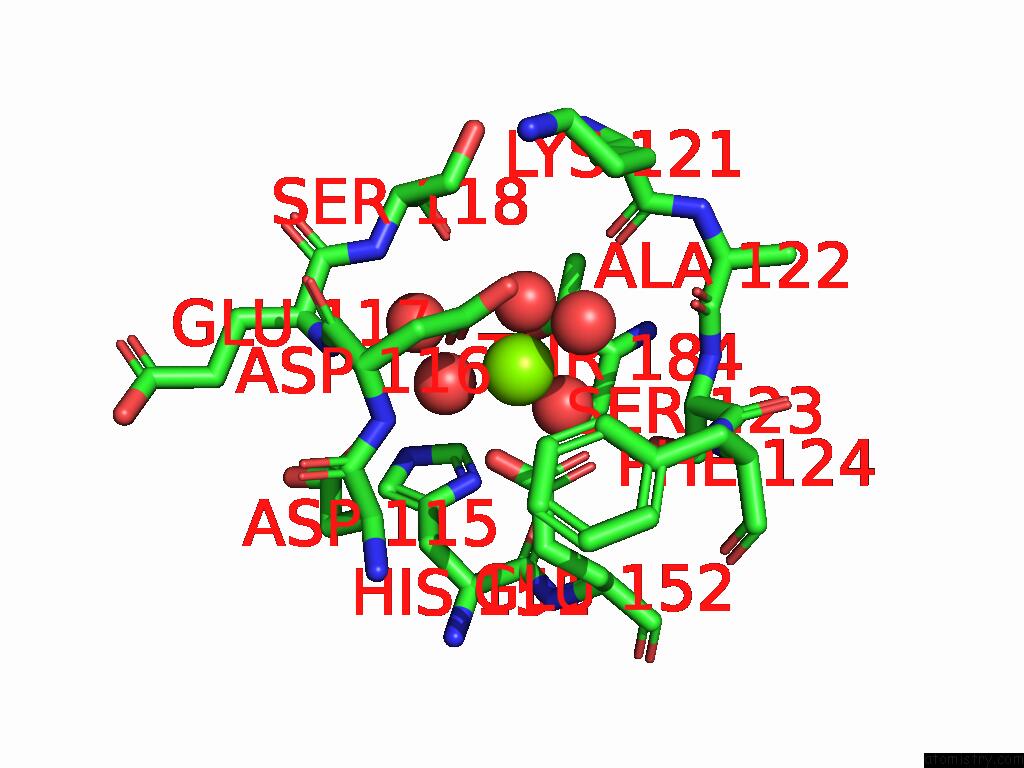
Mono view
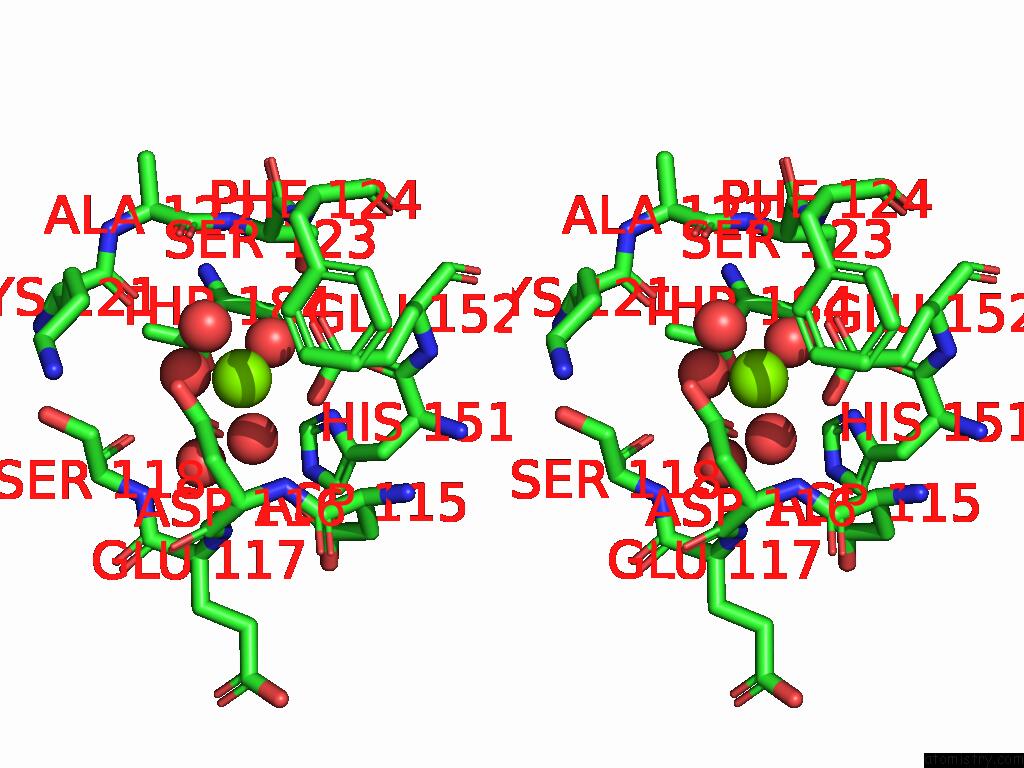
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Rhombotarget A within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 9cdv
Go back to
Magnesium binding site 3 out
of 3 in the Crystal Structure of Rhombotarget A
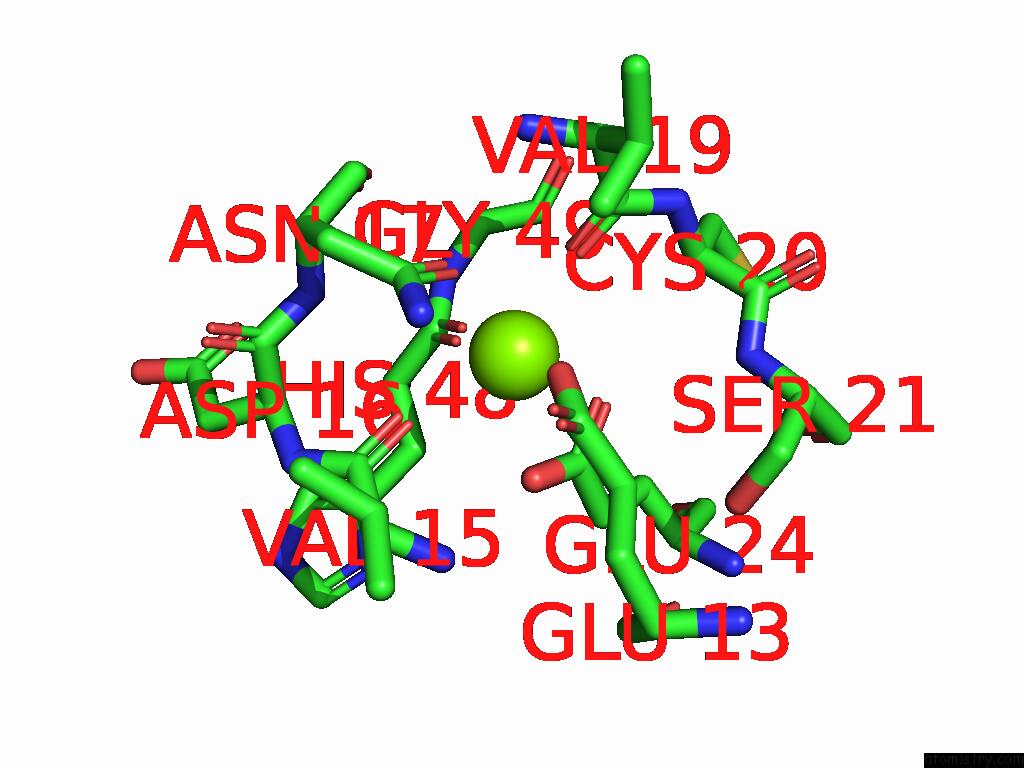
Mono view
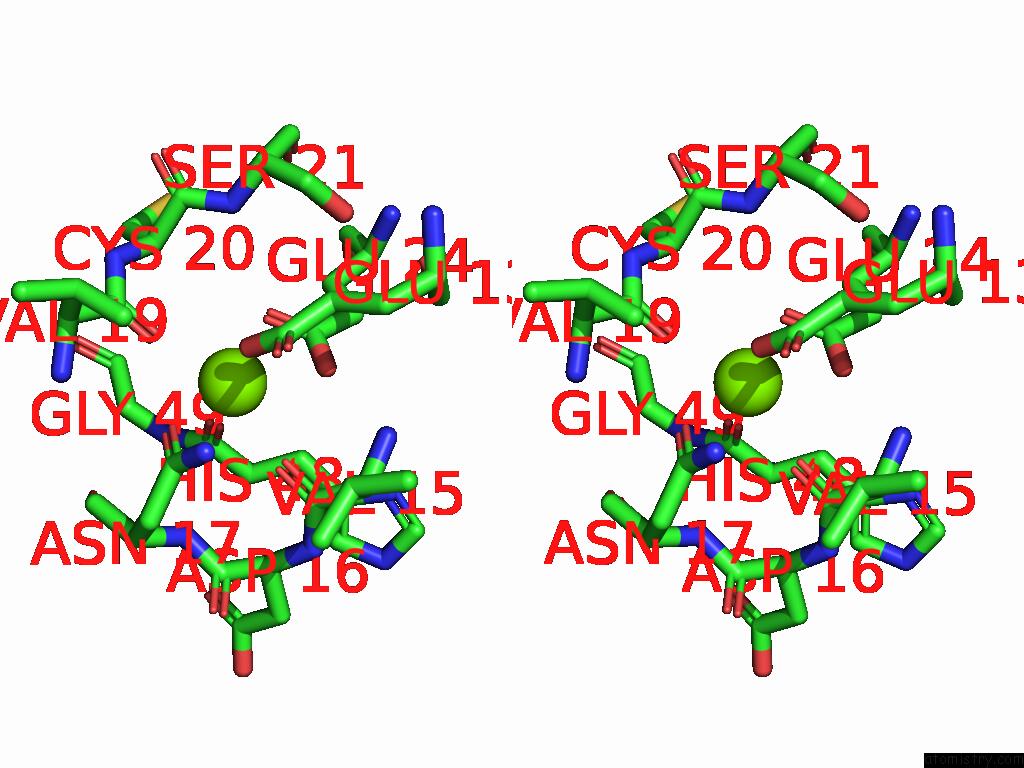
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of Rhombotarget A within 5.0Å range:
|
Reference:
S.A.Rettie,
D.Juergens,
V.Adebomi,
Y.F.Bueso,
Q.Zhao,
A.N.Leveille,
A.Liu,
A.K.Bera,
J.A.Wilms,
A.Uffing,
A.Kang,
E.Brackenbrough,
M.Lamb,
S.R.Gerben,
A.Murray,
P.M.Levine,
M.Schneider,
V.Vasireddy,
S.Ovchinnikov,
O.H.Weiergraber,
D.Willbold,
J.A.Kritzer,
J.D.Mougous,
D.Baker,
F.Dimaio,
G.Bhardwaj.
Accurate De Novo Design of High-Affinity Protein-Binding Macrocycles Using Deep Learning. Nat.Chem.Biol. 2025.
ISSN: ESSN 1552-4469
PubMed: 40542165
DOI: 10.1038/S41589-025-01929-W
Page generated: Sat Aug 16 00:08:38 2025
ISSN: ESSN 1552-4469
PubMed: 40542165
DOI: 10.1038/S41589-025-01929-W
Last articles
Na in 6BHDNa in 6BGK
Na in 6BG0
Na in 6BGQ
Na in 6BG1
Na in 6BAQ
Na in 6BFO
Na in 6BFK
Na in 6BEM
Na in 6BEL