Magnesium »
PDB 9cit-9d2b »
9cv1 »
Magnesium in PDB 9cv1: Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril
Enzymatic activity of Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril
All present enzymatic activity of Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril:
3.5.2.6;
3.5.2.6;
Protein crystallography data
The structure of Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril, PDB code: 9cv1
was solved by
S.B.Silwal,
J.C.Nix,
R.C.Page,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 39.63 / 1.29 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 67.75, 77.51, 79.26, 90, 90, 90 |
| R / Rfree (%) | 17.4 / 19.7 |
Other elements in 9cv1:
The structure of Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril
(pdb code 9cv1). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril, PDB code: 9cv1:
In total only one binding site of Magnesium was determined in the Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril, PDB code: 9cv1:
Magnesium binding site 1 out of 1 in 9cv1
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril
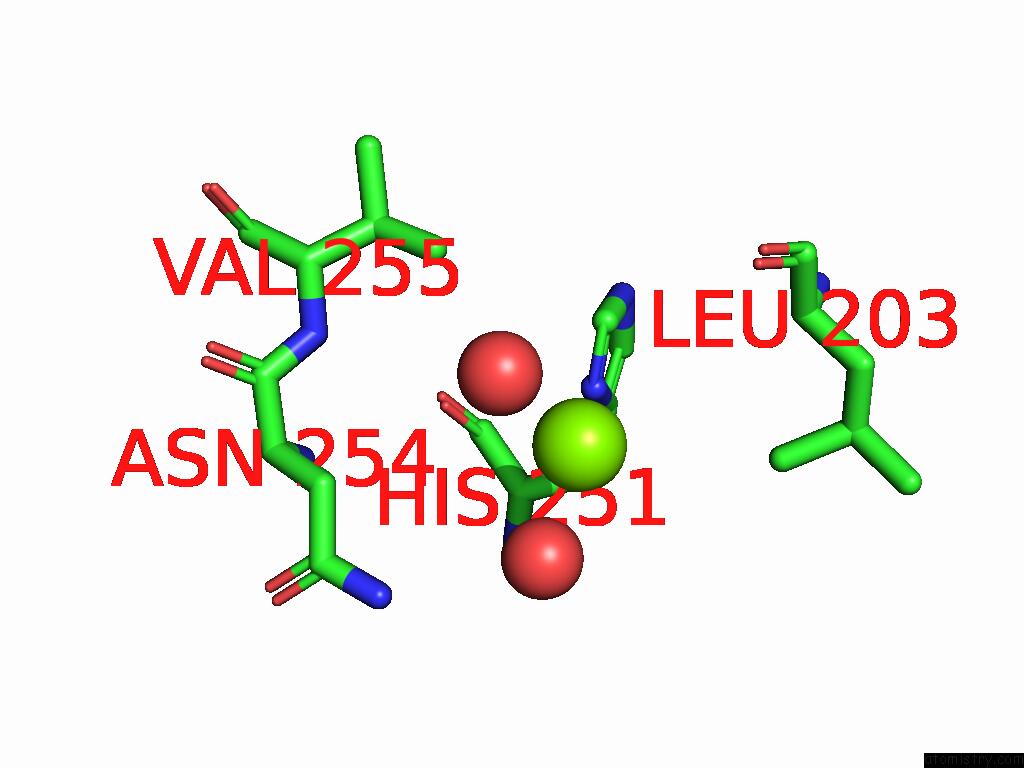
Mono view
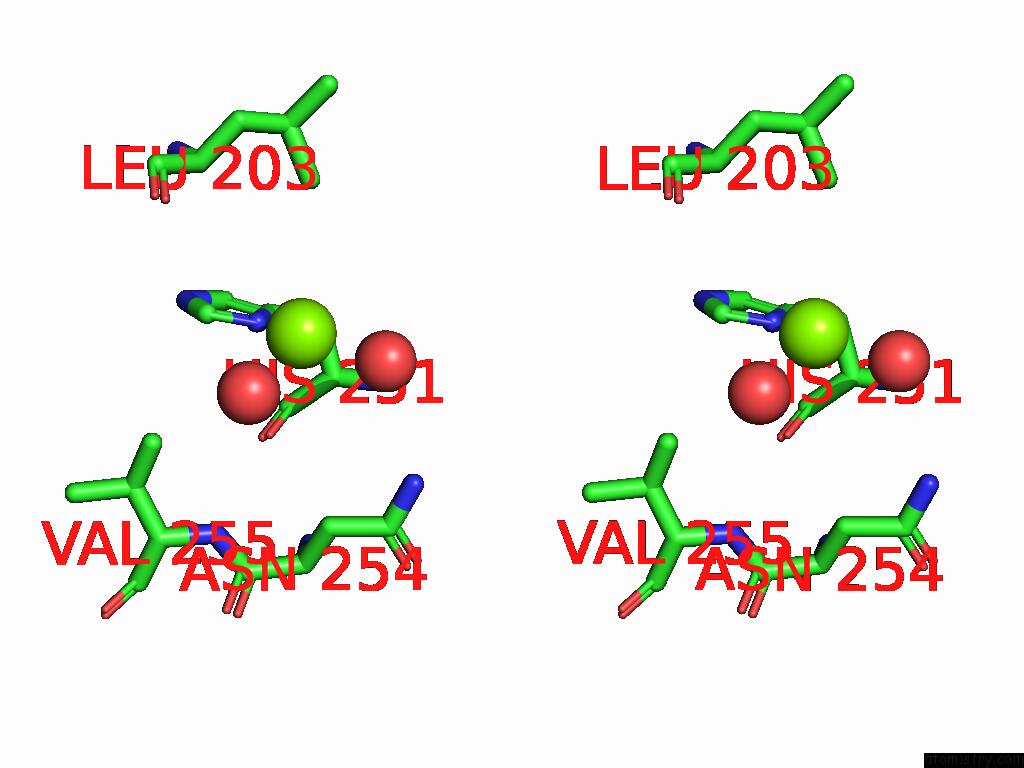
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Metallo-Beta-Lactamase Vim-15 with L- Captopril within 5.0Å range:
|
Reference:
S.B.Silwal,
B.Wamsley,
Z.Wang,
B.W.Gung,
J.C.Nix,
R.C.Page.
Mutation of An Active Site-Adjacent Residue in Vim Indirectly Dictates Interactions with and Blunts Inhibition By D-Captopril. J.Inorg.Biochem. V. 271 12975 2025.
ISSN: ISSN 0162-0134
PubMed: 40513263
DOI: 10.1016/J.JINORGBIO.2025.112975
Page generated: Sat Aug 16 00:18:36 2025
ISSN: ISSN 0162-0134
PubMed: 40513263
DOI: 10.1016/J.JINORGBIO.2025.112975
Last articles
Mn in 9LJUMn in 9LJW
Mn in 9LJS
Mn in 9LJR
Mn in 9LJT
Mn in 9LJV
Mg in 9UA2
Mg in 9R96
Mg in 9VM1
Mg in 9P01