Magnesium »
PDB 9jt2-9kqp »
9kev »
Magnesium in PDB 9kev: Cryo-Em Structure of Mycobacterium Tuberculosis Transcription Activation Complex with Six Phop Molecules (Composite Map)
Enzymatic activity of Cryo-Em Structure of Mycobacterium Tuberculosis Transcription Activation Complex with Six Phop Molecules (Composite Map)
All present enzymatic activity of Cryo-Em Structure of Mycobacterium Tuberculosis Transcription Activation Complex with Six Phop Molecules (Composite Map):
2.7.7.6;
2.7.7.6;
Other elements in 9kev:
The structure of Cryo-Em Structure of Mycobacterium Tuberculosis Transcription Activation Complex with Six Phop Molecules (Composite Map) also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of Mycobacterium Tuberculosis Transcription Activation Complex with Six Phop Molecules (Composite Map)
(pdb code 9kev). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of Mycobacterium Tuberculosis Transcription Activation Complex with Six Phop Molecules (Composite Map), PDB code: 9kev:
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of Mycobacterium Tuberculosis Transcription Activation Complex with Six Phop Molecules (Composite Map), PDB code: 9kev:
Magnesium binding site 1 out of 1 in 9kev
Go back to
Magnesium binding site 1 out
of 1 in the Cryo-Em Structure of Mycobacterium Tuberculosis Transcription Activation Complex with Six Phop Molecules (Composite Map)
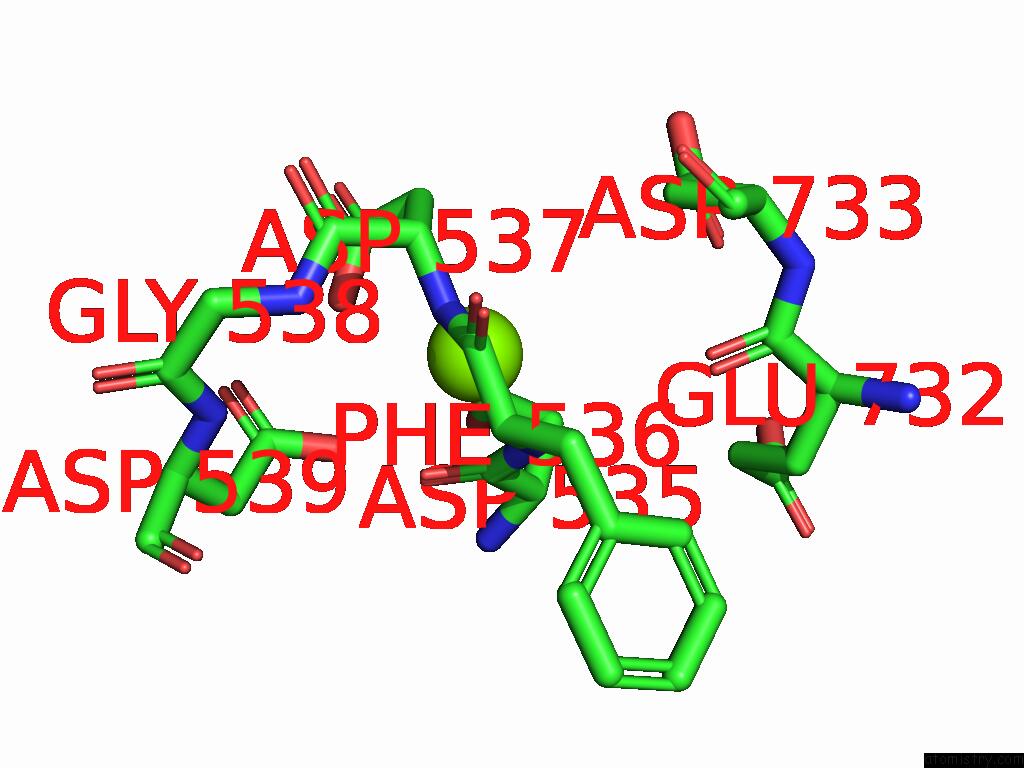
Mono view
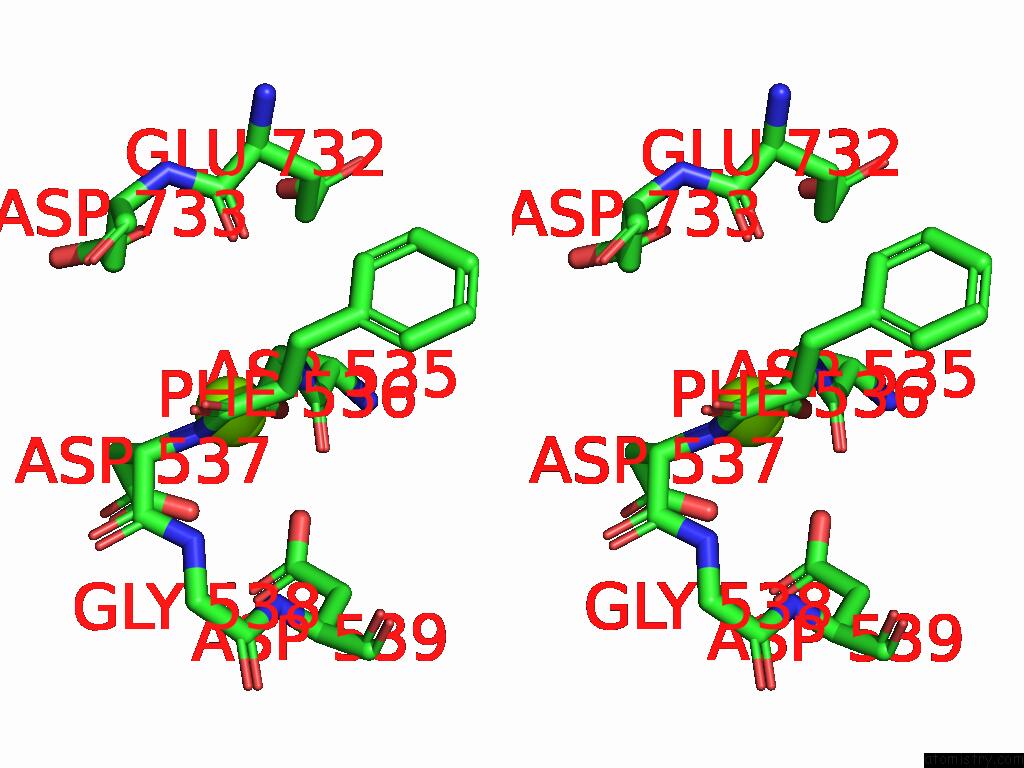
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of Mycobacterium Tuberculosis Transcription Activation Complex with Six Phop Molecules (Composite Map) within 5.0Å range:
|
Reference:
J.Shi,
Z.Feng,
Q.Song,
A.Wen,
T.Liu,
L.Xu,
Z.Ye,
S.Xu,
F.Gao,
L.Xiao,
J.Zhu,
K.Das,
G.Zhao,
J.Li,
Y.Feng,
W.Lin.
Structural Insights Into Transcription Regulation of the Global Ompr/Phob Family Regulator Phop From Mycobacterium Tuberculosis Nat Commun V. 16 1573 2025.
ISSN: ESSN 2041-1723
DOI: 10.1038/S41467-025-56697-X
Page generated: Sat Aug 16 04:56:20 2025
ISSN: ESSN 2041-1723
DOI: 10.1038/S41467-025-56697-X
Last articles
Mn in 2CYFMn in 2D0C
Mn in 2CY6
Mn in 2CWM
Mn in 2CTV
Mn in 2CM1
Mn in 2CPO
Mn in 2CNA
Mn in 2CJ2
Mn in 2CJ1