Magnesium »
PDB 9imj-9mm2 »
9mhg »
Magnesium in PDB 9mhg: Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation
Enzymatic activity of Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation
All present enzymatic activity of Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation:
2.7.1.137; 2.7.11.1; 3.6.5.2;
2.7.1.137; 2.7.11.1; 3.6.5.2;
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation
(pdb code 9mhg). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation, PDB code: 9mhg:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation, PDB code: 9mhg:
Jump to Magnesium binding site number: 1; 2;
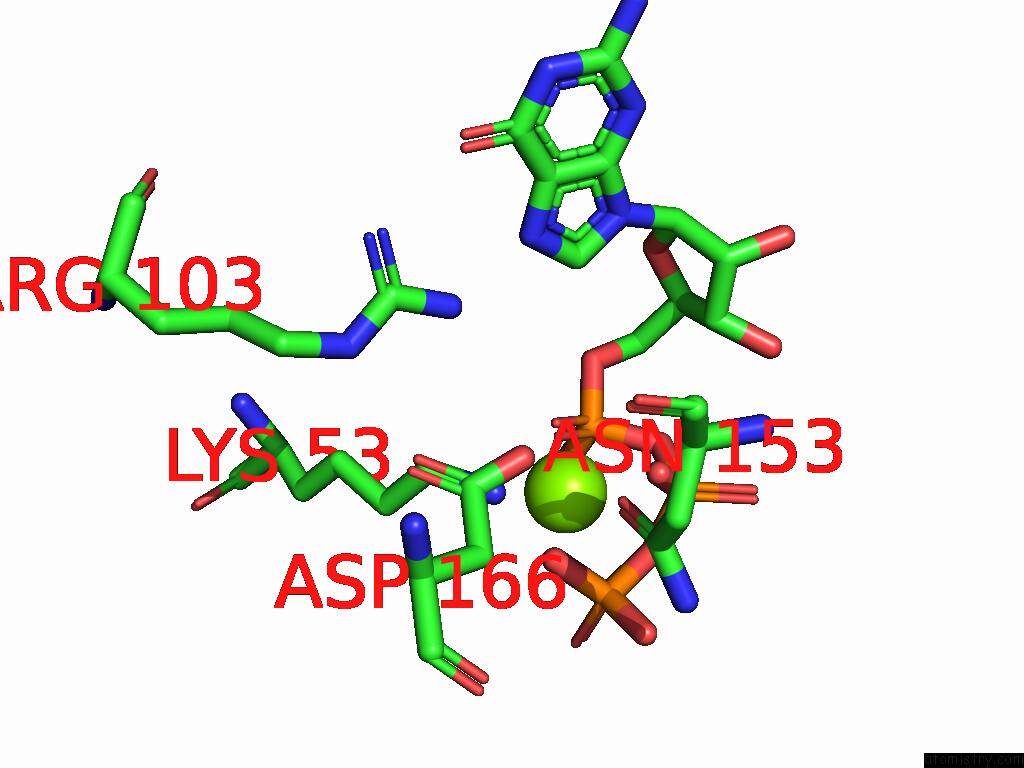
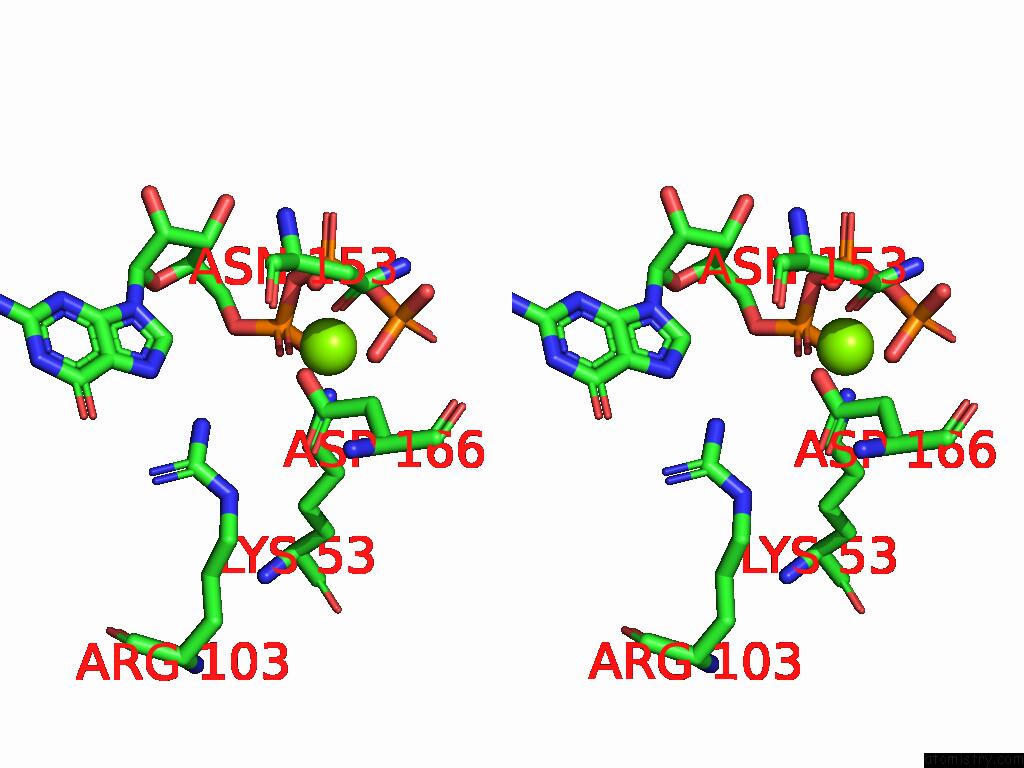
Magnesium binding site 1 out of 2 in 9mhg
Go back to
Magnesium binding site 1 out
of 2 in the Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation within 5.0Å range:
|
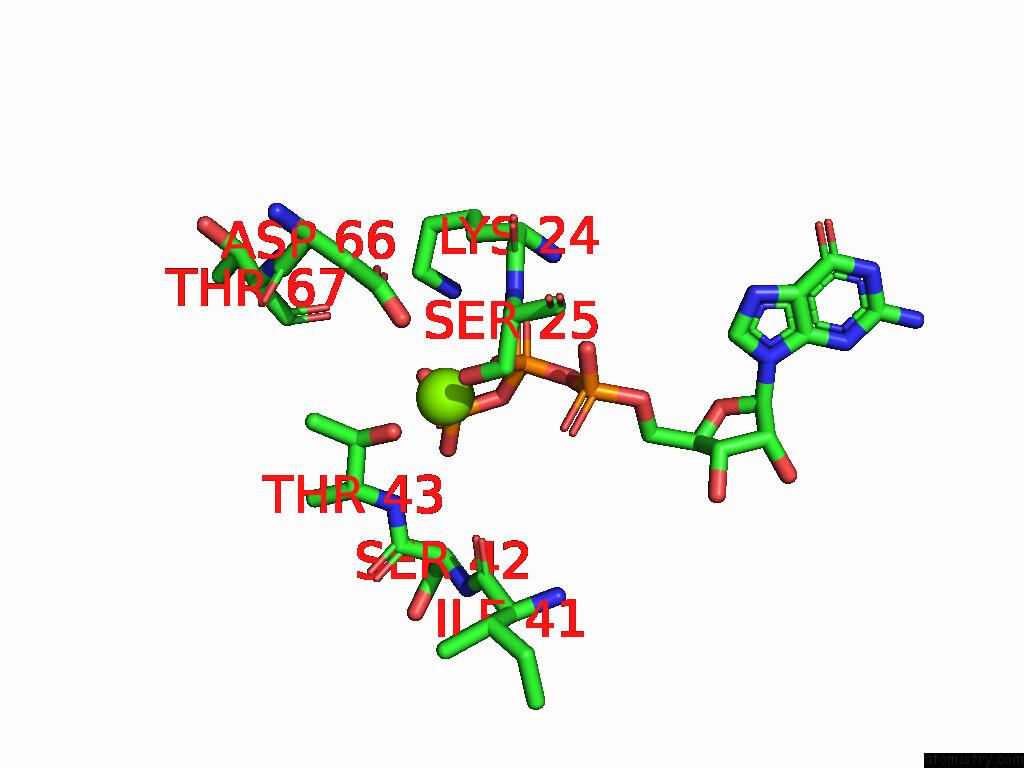
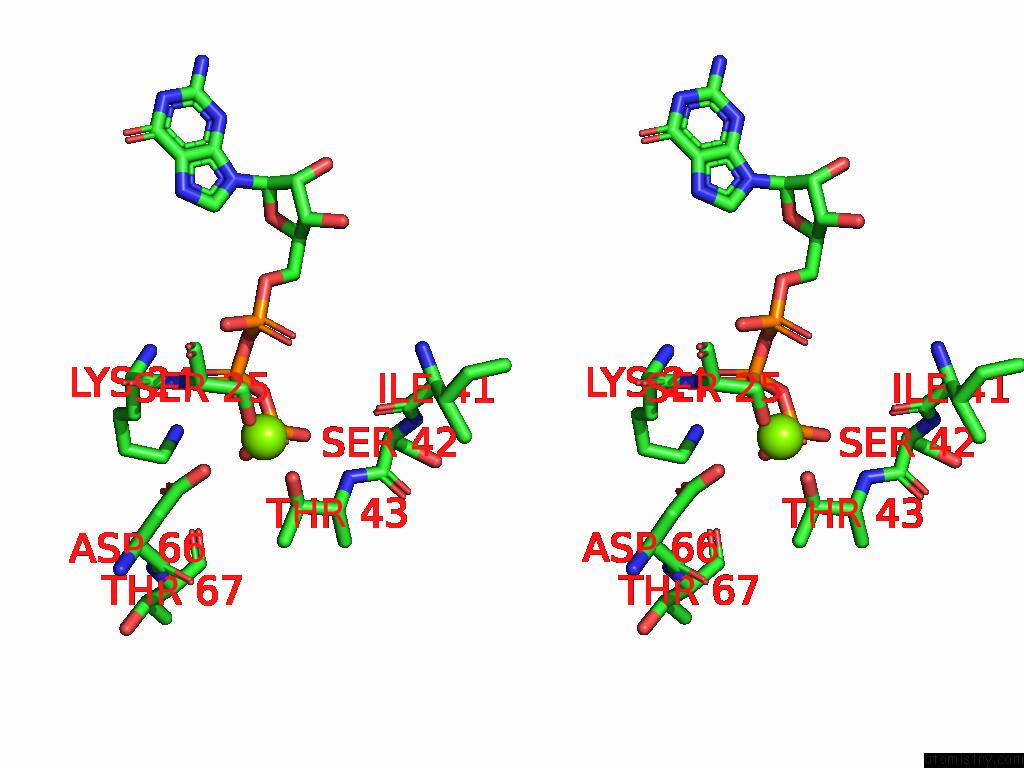
Magnesium binding site 2 out of 2 in 9mhg
Go back to
Magnesium binding site 2 out
of 2 in the Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Cryo Em Reconstruction of PI3KC3-C1 in Complex with Human RAB1A(Q70L), VPS34 Kinase Domain in the Inactive Conformation within 5.0Å range:
|
Reference:
A.S.I.Cook,
M.Chen,
T.N.Ngyuen,
A.Claveras-Cabezudo,
G.Khuu,
S.Rao,
S.N.Garcia,
M.Yang,
A.T.Iavarone,
X.Ren,
M.Lazarou,
G.Hummer,
J.H.Hurley.
Structural Pathway For Class III Pi 3-Kinase Activation By the Myristoylated Gtpbinding Pseudokinase VPS15 To Be Published.
Page generated: Tue Feb 25 11:23:02 2025
Last articles
F in 5FP5F in 5FHH
F in 5FI4
F in 5FHD
F in 5FI8
F in 5FHE
F in 5FHF
F in 5FDX
F in 5FEE
F in 5FD2