Magnesium »
PDB 1bho-1c5p »
1bl5 »
Magnesium in PDB 1bl5: Isocitrate Dehydrogenase From E. Coli Single Turnover Laue Structure of Rate-Limited Product Complex, 10 Msec Time Resolution
Enzymatic activity of Isocitrate Dehydrogenase From E. Coli Single Turnover Laue Structure of Rate-Limited Product Complex, 10 Msec Time Resolution
All present enzymatic activity of Isocitrate Dehydrogenase From E. Coli Single Turnover Laue Structure of Rate-Limited Product Complex, 10 Msec Time Resolution:
1.1.1.42;
1.1.1.42;
Protein crystallography data
The structure of Isocitrate Dehydrogenase From E. Coli Single Turnover Laue Structure of Rate-Limited Product Complex, 10 Msec Time Resolution, PDB code: 1bl5
was solved by
B.L.Stoddard,
B.Cohen,
M.Brubaker,
A.Mesecar,
D.E.Koshland Junior,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.50 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 106.100, 106.100, 151.800, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.2 / 27.1 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Isocitrate Dehydrogenase From E. Coli Single Turnover Laue Structure of Rate-Limited Product Complex, 10 Msec Time Resolution
(pdb code 1bl5). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Isocitrate Dehydrogenase From E. Coli Single Turnover Laue Structure of Rate-Limited Product Complex, 10 Msec Time Resolution, PDB code: 1bl5:
In total only one binding site of Magnesium was determined in the Isocitrate Dehydrogenase From E. Coli Single Turnover Laue Structure of Rate-Limited Product Complex, 10 Msec Time Resolution, PDB code: 1bl5:
Magnesium binding site 1 out of 1 in 1bl5
Go back to
Magnesium binding site 1 out
of 1 in the Isocitrate Dehydrogenase From E. Coli Single Turnover Laue Structure of Rate-Limited Product Complex, 10 Msec Time Resolution
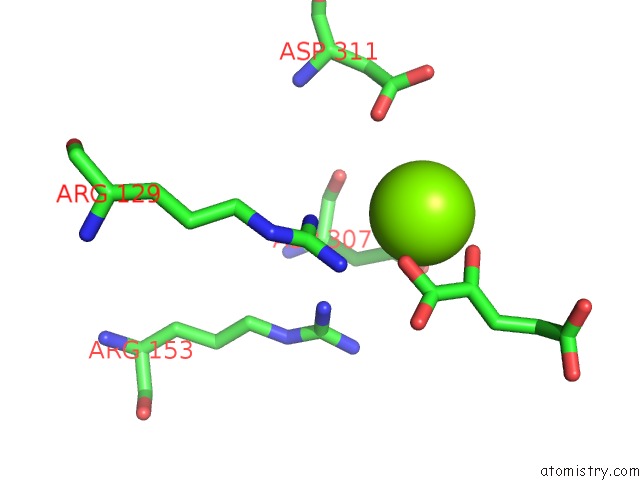
Mono view
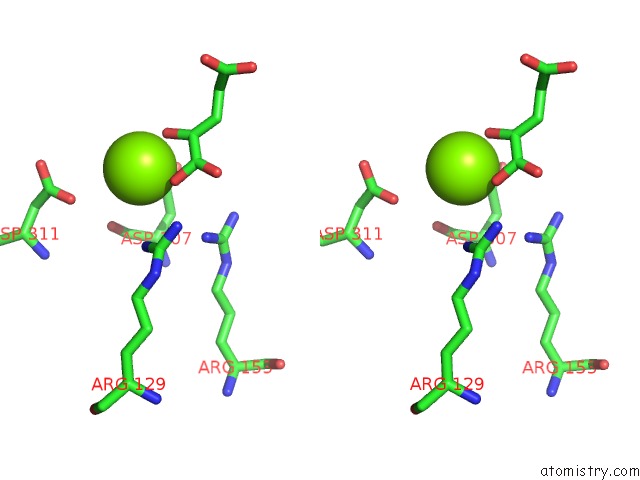
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Isocitrate Dehydrogenase From E. Coli Single Turnover Laue Structure of Rate-Limited Product Complex, 10 Msec Time Resolution within 5.0Å range:
|
Reference:
B.L.Stoddard,
B.E.Cohen,
M.Brubaker,
A.D.Mesecar,
D.E.Koshland Jr..
Millisecond Laue Structures of An Enzyme-Product Complex Using Photocaged Substrate Analogs. Nat.Struct.Biol. V. 5 891 1998.
ISSN: ISSN 1072-8368
PubMed: 9783749
DOI: 10.1038/2331
Page generated: Sat Aug 9 20:12:24 2025
ISSN: ISSN 1072-8368
PubMed: 9783749
DOI: 10.1038/2331
Last articles
Mg in 2DFTMg in 2DKG
Mg in 2DH4
Mg in 2DGN
Mg in 2DES
Mg in 2DDT
Mg in 2DEI
Mg in 2DEJ
Mg in 2DE3
Mg in 2DDX