Magnesium »
PDB 2hld-2hxf »
2hxf »
Magnesium in PDB 2hxf: KIF1A Head-Microtubule Complex Structure in Amppnp-Form
Magnesium Binding Sites:
The binding sites of Magnesium atom in the KIF1A Head-Microtubule Complex Structure in Amppnp-Form
(pdb code 2hxf). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the KIF1A Head-Microtubule Complex Structure in Amppnp-Form, PDB code: 2hxf:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the KIF1A Head-Microtubule Complex Structure in Amppnp-Form, PDB code: 2hxf:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 2hxf
Go back to
Magnesium binding site 1 out
of 2 in the KIF1A Head-Microtubule Complex Structure in Amppnp-Form
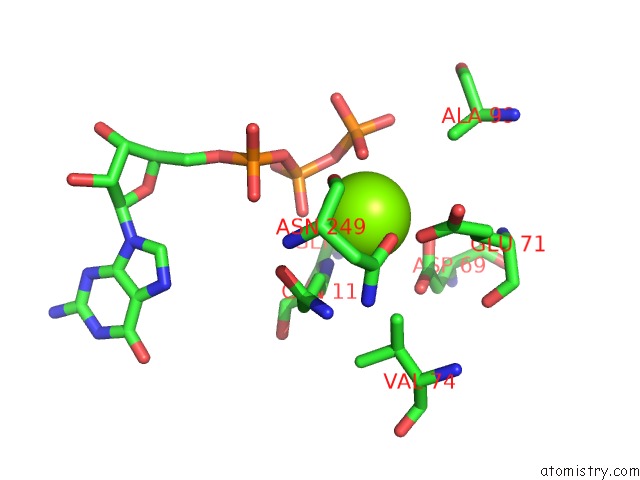
Mono view
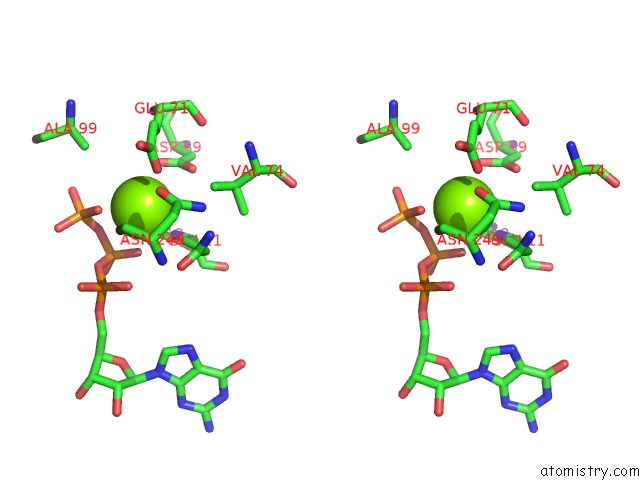
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of KIF1A Head-Microtubule Complex Structure in Amppnp-Form within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 2hxf
Go back to
Magnesium binding site 2 out
of 2 in the KIF1A Head-Microtubule Complex Structure in Amppnp-Form
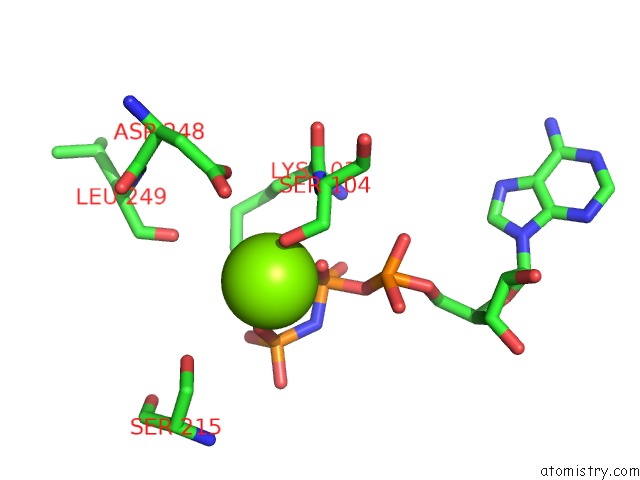
Mono view
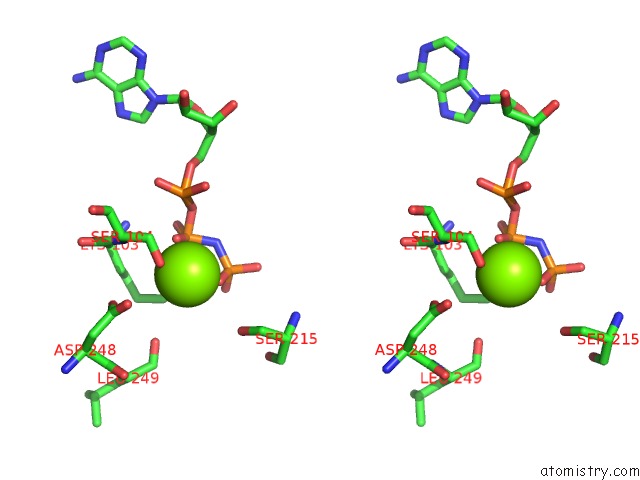
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of KIF1A Head-Microtubule Complex Structure in Amppnp-Form within 5.0Å range:
|
Reference:
M.Kikkawa,
N.Hirokawa.
High-Resolution Cryo-Em Maps Show the Nucleotide Binding Pocket of KIF1A in Open and Closed Conformations Embo J. V. 25 4187 2006.
ISSN: ISSN 0261-4189
PubMed: 16946706
DOI: 10.1038/SJ.EMBOJ.7601299
Page generated: Sun Aug 10 11:29:33 2025
ISSN: ISSN 0261-4189
PubMed: 16946706
DOI: 10.1038/SJ.EMBOJ.7601299
Last articles
Mg in 5XP0Mg in 5XOU
Mg in 5XOW
Mg in 5XON
Mg in 5XOJ
Mg in 5XOG
Mg in 5XOB
Mg in 5XNN
Mg in 5XNX
Mg in 5XNO