Magnesium »
PDB 2xbp-2xnd »
2xjc »
Magnesium in PDB 2xjc: Crystal Structure of the D52N Variant of Cytosolic 5'-Nucleotidase II in Complex with Guanosine Monophosphate and Diadenosine Tetraphosphate
Enzymatic activity of Crystal Structure of the D52N Variant of Cytosolic 5'-Nucleotidase II in Complex with Guanosine Monophosphate and Diadenosine Tetraphosphate
All present enzymatic activity of Crystal Structure of the D52N Variant of Cytosolic 5'-Nucleotidase II in Complex with Guanosine Monophosphate and Diadenosine Tetraphosphate:
3.1.3.5;
3.1.3.5;
Protein crystallography data
The structure of Crystal Structure of the D52N Variant of Cytosolic 5'-Nucleotidase II in Complex with Guanosine Monophosphate and Diadenosine Tetraphosphate, PDB code: 2xjc
was solved by
K.Wallden,
P.Nordlund,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.79 / 2.00 |
| Space group | I 2 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 91.540, 127.360, 130.240, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.58 / 21.893 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the D52N Variant of Cytosolic 5'-Nucleotidase II in Complex with Guanosine Monophosphate and Diadenosine Tetraphosphate
(pdb code 2xjc). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of the D52N Variant of Cytosolic 5'-Nucleotidase II in Complex with Guanosine Monophosphate and Diadenosine Tetraphosphate, PDB code: 2xjc:
In total only one binding site of Magnesium was determined in the Crystal Structure of the D52N Variant of Cytosolic 5'-Nucleotidase II in Complex with Guanosine Monophosphate and Diadenosine Tetraphosphate, PDB code: 2xjc:
Magnesium binding site 1 out of 1 in 2xjc
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of the D52N Variant of Cytosolic 5'-Nucleotidase II in Complex with Guanosine Monophosphate and Diadenosine Tetraphosphate
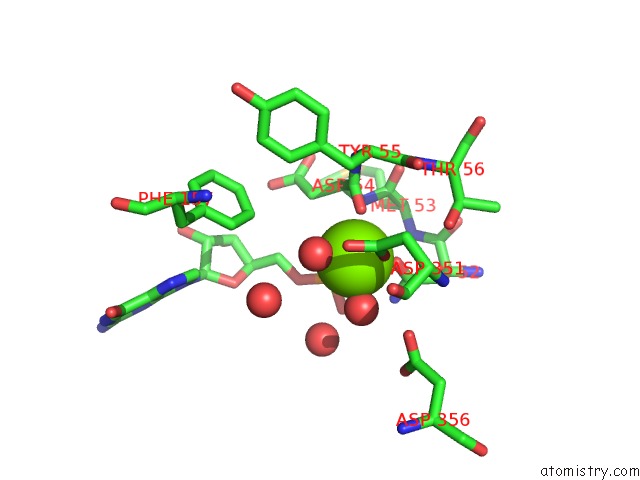
Mono view
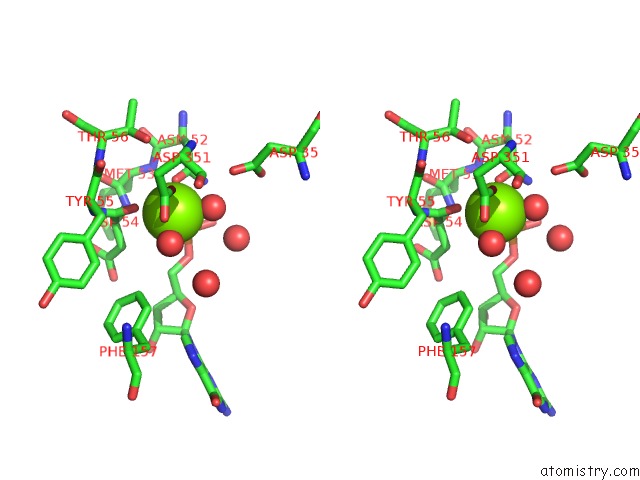
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the D52N Variant of Cytosolic 5'-Nucleotidase II in Complex with Guanosine Monophosphate and Diadenosine Tetraphosphate within 5.0Å range:
|
Reference:
K.Wallden,
P.Nordlund.
Structural Basis For the Allosteric Regulation and Substrate Recognition of Human Cytosolic 5'-Nucleotidase II J.Mol.Biol. V. 408 684 2011.
ISSN: ISSN 0022-2836
PubMed: 21396942
DOI: 10.1016/J.JMB.2011.02.059
Page generated: Sun Aug 10 16:27:40 2025
ISSN: ISSN 0022-2836
PubMed: 21396942
DOI: 10.1016/J.JMB.2011.02.059
Last articles
Mg in 5ZKJMg in 5ZKI
Mg in 5ZK6
Mg in 5ZE9
Mg in 5ZFX
Mg in 5ZCT
Mg in 5ZE6
Mg in 5ZE4
Mg in 5ZDN
Mg in 5ZE0