Magnesium »
PDB 3bdh-3bre »
3bn3 »
Magnesium in PDB 3bn3: Crystal Structure of Icam-5 in Complex with Al I Domain
Protein crystallography data
The structure of Crystal Structure of Icam-5 in Complex with Al I Domain, PDB code: 3bn3
was solved by
H.Zhang,
T.A.Springer,
J.-H.Wang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 71.98 / 2.10 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 61.793, 71.507, 143.871, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 19.1 / 23.5 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Icam-5 in Complex with Al I Domain
(pdb code 3bn3). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Icam-5 in Complex with Al I Domain, PDB code: 3bn3:
In total only one binding site of Magnesium was determined in the Crystal Structure of Icam-5 in Complex with Al I Domain, PDB code: 3bn3:
Magnesium binding site 1 out of 1 in 3bn3
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Icam-5 in Complex with Al I Domain
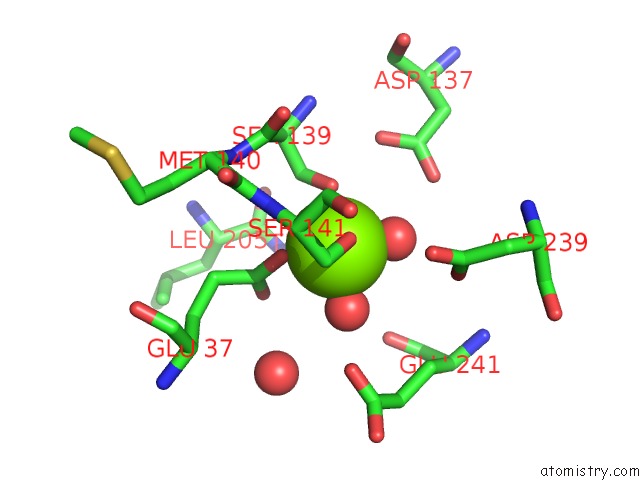
Mono view
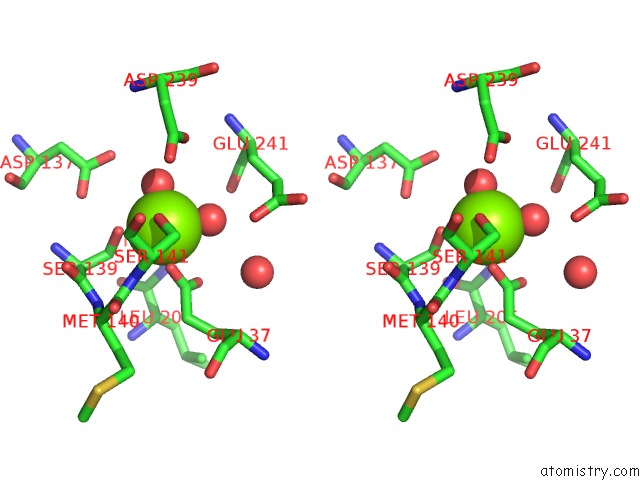
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Icam-5 in Complex with Al I Domain within 5.0Å range:
|
Reference:
H.Zhang,
J.M.Casasnovas,
M.Jin,
J.H.Liu,
C.G.Gahmberg,
T.A.Springer,
J.-H.Wang.
An Unusual Allosteric Mobility of the C-Terminal Helix of A High-Affinity Alpha L Integrin I Domain Variant Bound to Icam-5 Mol.Cell V. 31 432 2008.
ISSN: ISSN 1097-2765
PubMed: 18691975
DOI: 10.1016/J.MOLCEL.2008.06.022
Page generated: Sun Aug 10 17:51:00 2025
ISSN: ISSN 1097-2765
PubMed: 18691975
DOI: 10.1016/J.MOLCEL.2008.06.022
Last articles
Mg in 5Y4NMg in 5Y4J
Mg in 5Y4I
Mg in 5Y3J
Mg in 5Y41
Mg in 5Y3I
Mg in 5Y36
Mg in 5Y34
Mg in 5Y1W
Mg in 5Y1X