Magnesium »
PDB 3nl0-3o0q »
3nyr »
Magnesium in PDB 3nyr: Malonyl-Coa Ligase Ternary Product Complex with Malonyl-Coa and Amp Bound
Protein crystallography data
The structure of Malonyl-Coa Ligase Ternary Product Complex with Malonyl-Coa and Amp Bound, PDB code: 3nyr
was solved by
A.J.Hughes,
A.T.Keatinge-Clay,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 76.57 / 1.45 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 73.191, 86.628, 153.149, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20 / 22.5 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Malonyl-Coa Ligase Ternary Product Complex with Malonyl-Coa and Amp Bound
(pdb code 3nyr). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Malonyl-Coa Ligase Ternary Product Complex with Malonyl-Coa and Amp Bound, PDB code: 3nyr:
In total only one binding site of Magnesium was determined in the Malonyl-Coa Ligase Ternary Product Complex with Malonyl-Coa and Amp Bound, PDB code: 3nyr:
Magnesium binding site 1 out of 1 in 3nyr
Go back to
Magnesium binding site 1 out
of 1 in the Malonyl-Coa Ligase Ternary Product Complex with Malonyl-Coa and Amp Bound
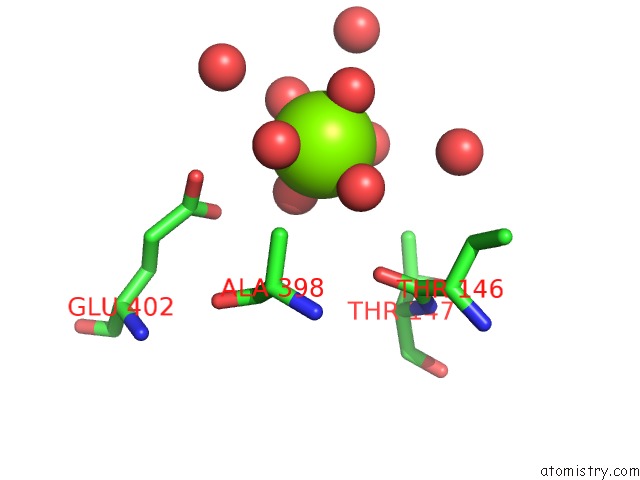
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Malonyl-Coa Ligase Ternary Product Complex with Malonyl-Coa and Amp Bound within 5.0Å range:
|
Reference:
A.J.Hughes,
A.Keatinge-Clay.
Enzymatic Extender Unit Generation For in Vitro Polyketide Synthase Reactions: Structural and Functional Showcasing of Streptomyces Coelicolor Matb. Chem.Biol. V. 18 165 2011.
ISSN: ISSN 1074-5521
PubMed: 21338915
DOI: 10.1016/J.CHEMBIOL.2010.12.014
Page generated: Mon Aug 11 01:01:19 2025
ISSN: ISSN 1074-5521
PubMed: 21338915
DOI: 10.1016/J.CHEMBIOL.2010.12.014
Last articles
Mg in 6KPEMg in 6KOQ
Mg in 6KNZ
Mg in 6KOP
Mg in 6KON
Mg in 6KOO
Mg in 6KO6
Mg in 6KO1
Mg in 6KMA
Mg in 6KO0