Magnesium »
PDB 3tav-3tnf »
3tdw »
Magnesium in PDB 3tdw: The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant
Protein crystallography data
The structure of The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant, PDB code: 3tdw
was solved by
C.A.Smith,
S.B.Vakulenko,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.21 / 1.70 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 83.060, 54.690, 77.810, 90.00, 108.49, 90.00 |
| R / Rfree (%) | 17.4 / 22 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant
(pdb code 3tdw). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant, PDB code: 3tdw:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant, PDB code: 3tdw:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 3tdw
Go back to
Magnesium binding site 1 out
of 4 in the The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant within 5.0Å range:
|
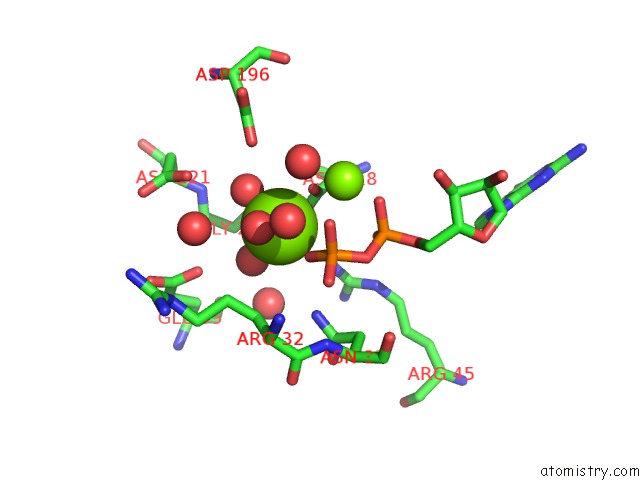
Magnesium binding site 2 out of 4 in 3tdw
Go back to
Magnesium binding site 2 out
of 4 in the The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant within 5.0Å range:
|
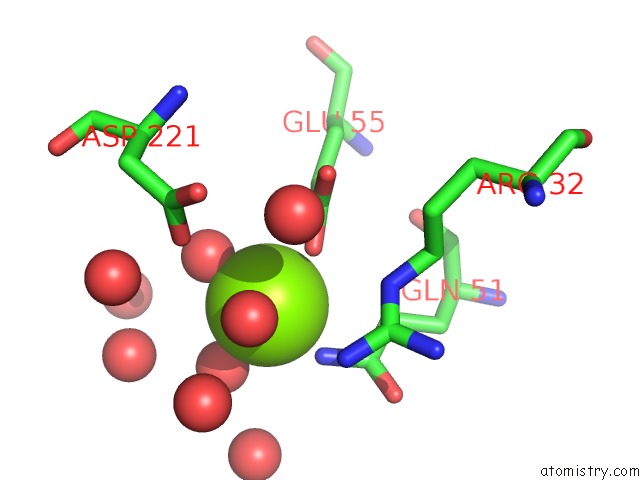
Magnesium binding site 3 out of 4 in 3tdw
Go back to
Magnesium binding site 3 out
of 4 in the The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 3tdw
Go back to
Magnesium binding site 4 out
of 4 in the The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of The Gdp Complex of the Aminoglycoside 2'-Phosphotransfere-Iiia F108L Mutant within 5.0Å range:
|
Reference:
C.A.Smith,
M.Toth,
H.Frase,
L.J.Byrnes,
S.B.Vakulenko.
Aminoglycoside 2''-Phosphotransferase Iiia (Aph(2'')-Iiia) Prefers Gtp Over Atp: Structural Templates For Nucleotide Recognition in the Bacterial Aminoglycoside-2'' Kinases. J.Biol.Chem. V. 287 12893 2012.
ISSN: ISSN 0021-9258
PubMed: 22367198
DOI: 10.1074/JBC.M112.341206
Page generated: Mon Aug 11 03:53:21 2025
ISSN: ISSN 0021-9258
PubMed: 22367198
DOI: 10.1074/JBC.M112.341206
Last articles
Mg in 4AP5Mg in 4AOU
Mg in 4ANJ
Mg in 4AOV
Mg in 4AN9
Mg in 4AN2
Mg in 4ANB
Mg in 4AN3
Mg in 4ALX
Mg in 4AMS