Magnesium »
PDB 4ix4-4j9k »
4j0q »
Magnesium in PDB 4j0q: Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu)
Protein crystallography data
The structure of Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu), PDB code: 4j0q
was solved by
J.S.Scotti,
M.A.Mcdonough,
C.J.Schofield,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.24 / 2.29 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 212.505, 155.624, 99.298, 90.00, 114.76, 90.00 |
| R / Rfree (%) | 17.4 / 21.3 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu)
(pdb code 4j0q). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 5 binding sites of Magnesium where determined in the Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu), PDB code: 4j0q:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Magnesium where determined in the Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu), PDB code: 4j0q:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5;
Magnesium binding site 1 out of 5 in 4j0q
Go back to
Magnesium binding site 1 out
of 5 in the Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu)
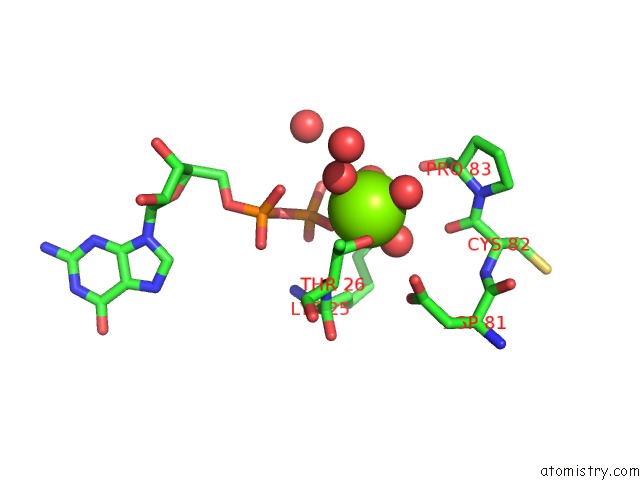
Mono view
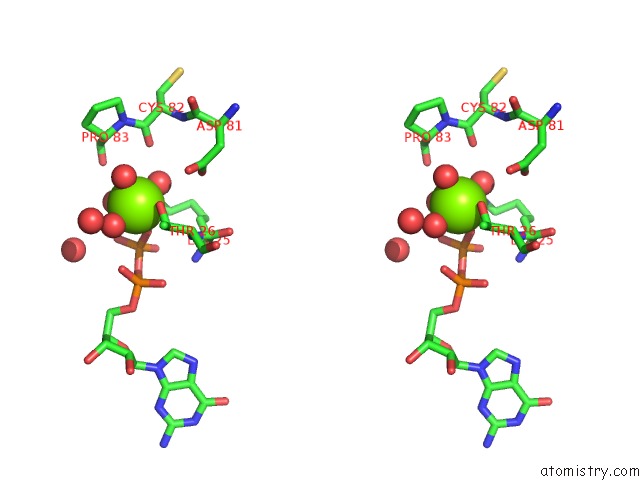
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu) within 5.0Å range:
|
Magnesium binding site 2 out of 5 in 4j0q
Go back to
Magnesium binding site 2 out
of 5 in the Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu)
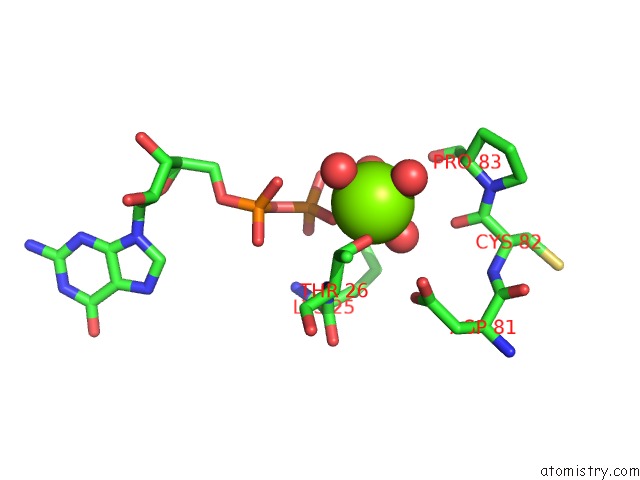
Mono view
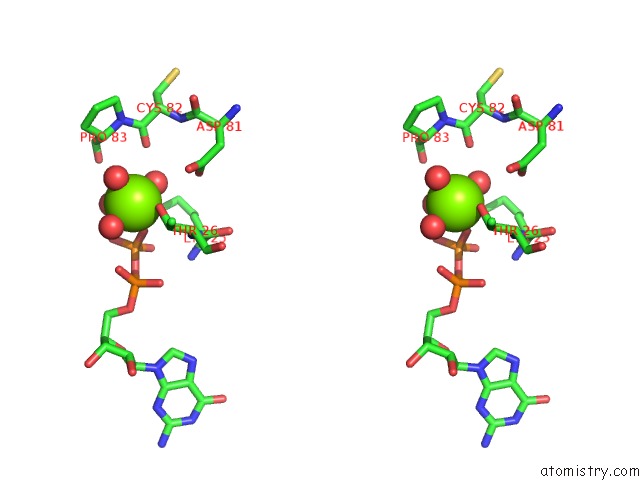
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu) within 5.0Å range:
|
Magnesium binding site 3 out of 5 in 4j0q
Go back to
Magnesium binding site 3 out
of 5 in the Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu)
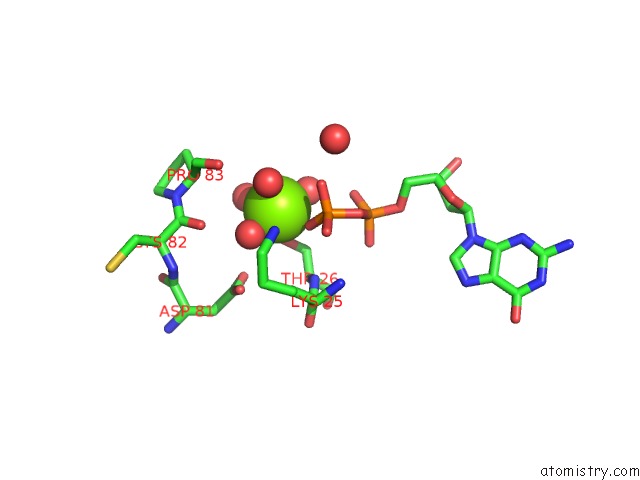
Mono view
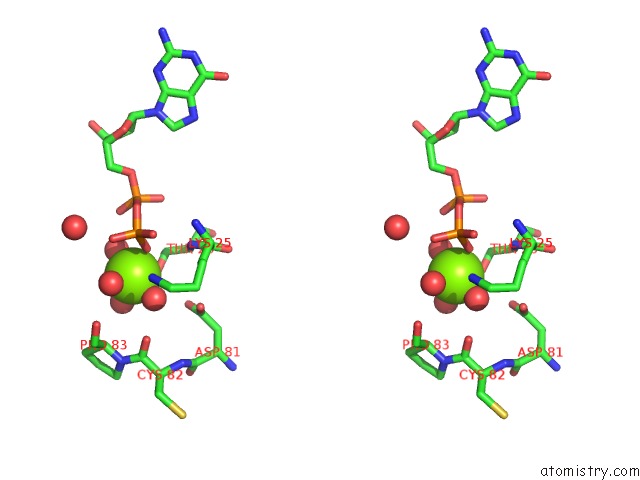
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu) within 5.0Å range:
|
Magnesium binding site 4 out of 5 in 4j0q
Go back to
Magnesium binding site 4 out
of 5 in the Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu)
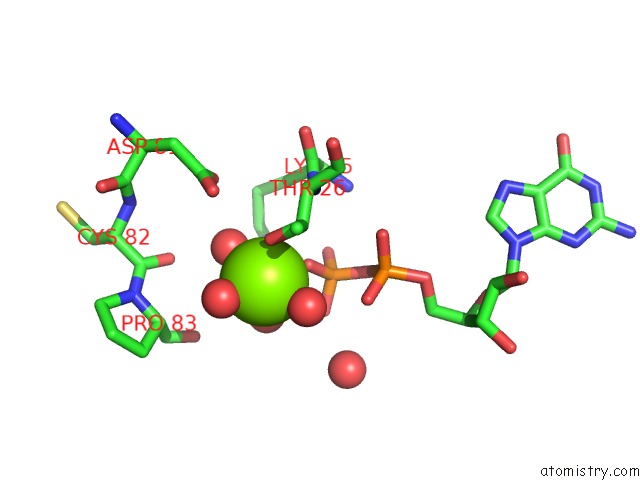
Mono view
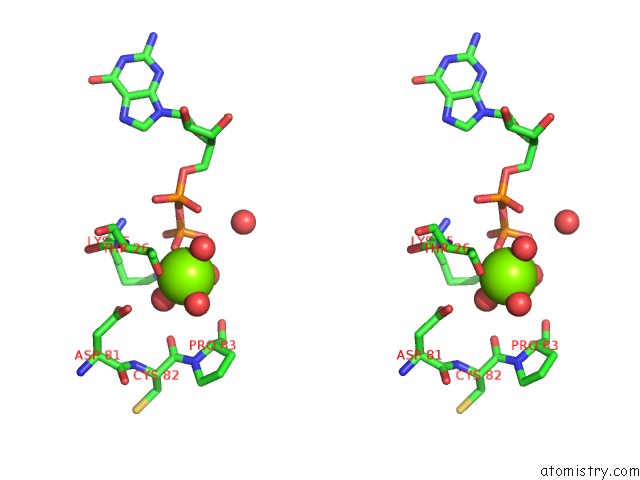
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu) within 5.0Å range:
|
Magnesium binding site 5 out of 5 in 4j0q
Go back to
Magnesium binding site 5 out
of 5 in the Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu)
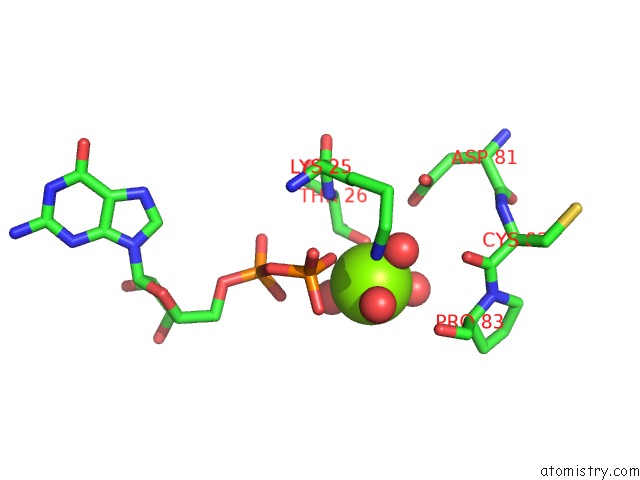
Mono view
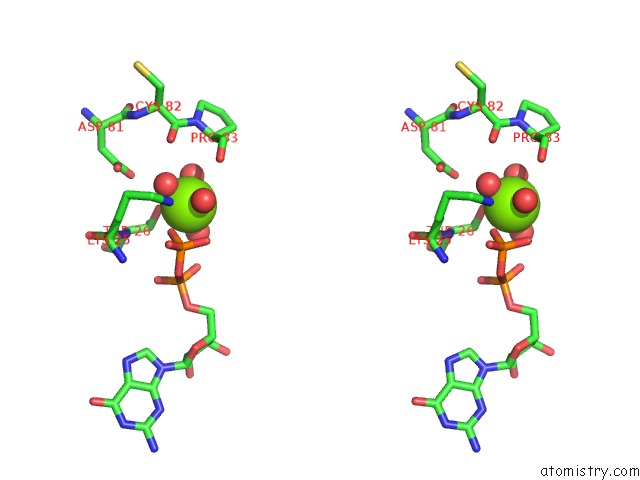
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of Pseudomonas Putida Elongation Factor Tu (Ef-Tu) within 5.0Å range:
|
Reference:
J.S.Scotti,
I.K.H.Leung,
W.Ge,
M.A.Bentley,
J.Paps,
H.B.Kramer,
J.Lee,
W.Aik,
H.Choi,
S.M.Paulsen,
L.A.H.Bowman,
N.D.Loik,
S.Horita,
C.H.Ho,
N.J.Kershaw,
C.M.Tang,
T.D.W.Claridge,
G.M.Preston,
M.A.Mcdonough,
C.J.Schofield.
Human Oxygen Sensing May Have Origins in Prokaryotic Elongation Factor Tu Prolyl-Hydroxylation Proc.Natl.Acad.Sci.Usa V. 111 13331 2014.
ISSN: ISSN 0027-8424
PubMed: 25197067
DOI: 10.1073/PNAS.1409916111
Page generated: Mon Aug 11 14:30:55 2025
ISSN: ISSN 0027-8424
PubMed: 25197067
DOI: 10.1073/PNAS.1409916111
Last articles
Mg in 4LGYMg in 4LHU
Mg in 4LH5
Mg in 4LH4
Mg in 4LGD
Mg in 4LGB
Mg in 4LGA
Mg in 4LG5
Mg in 4LFG
Mg in 4LFV