Magnesium »
PDB 4j9l-4jjs »
4jej »
Magnesium in PDB 4jej: Gggps From Flavobacterium Johnsoniae
Protein crystallography data
The structure of Gggps From Flavobacterium Johnsoniae, PDB code: 4jej
was solved by
D.Peterhoff,
B.Beer,
C.Rajendran,
E.P.Kumpula,
E.Kapetaniou,
H.Guldan,
R.K.Wierenga,
R.Sterner,
P.Babinger,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.06 / 1.52 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 82.270, 43.010, 75.927, 90.00, 117.41, 90.00 |
| R / Rfree (%) | 17.7 / 19.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Gggps From Flavobacterium Johnsoniae
(pdb code 4jej). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Gggps From Flavobacterium Johnsoniae, PDB code: 4jej:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Gggps From Flavobacterium Johnsoniae, PDB code: 4jej:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 4jej
Go back to
Magnesium binding site 1 out
of 2 in the Gggps From Flavobacterium Johnsoniae
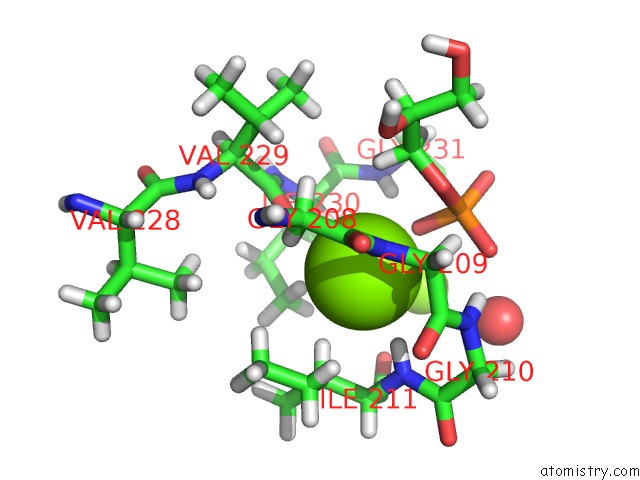
Mono view
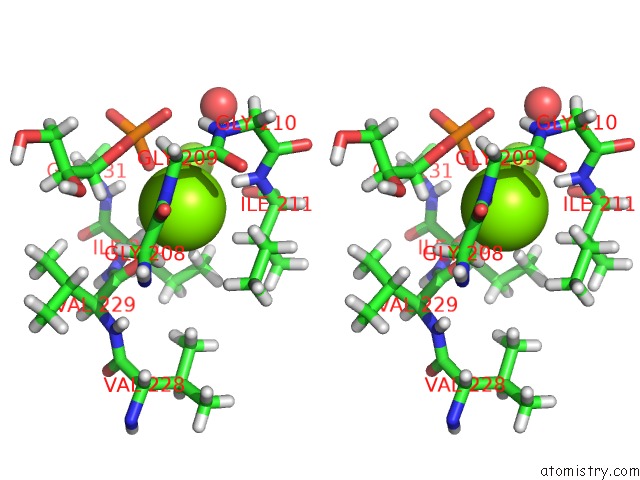
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Gggps From Flavobacterium Johnsoniae within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 4jej
Go back to
Magnesium binding site 2 out
of 2 in the Gggps From Flavobacterium Johnsoniae
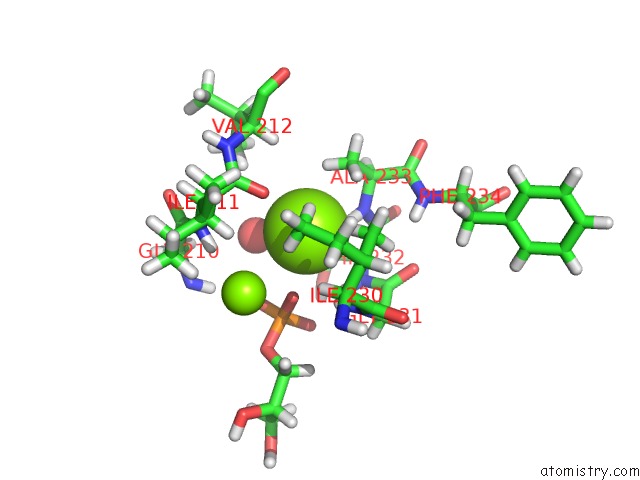
Mono view
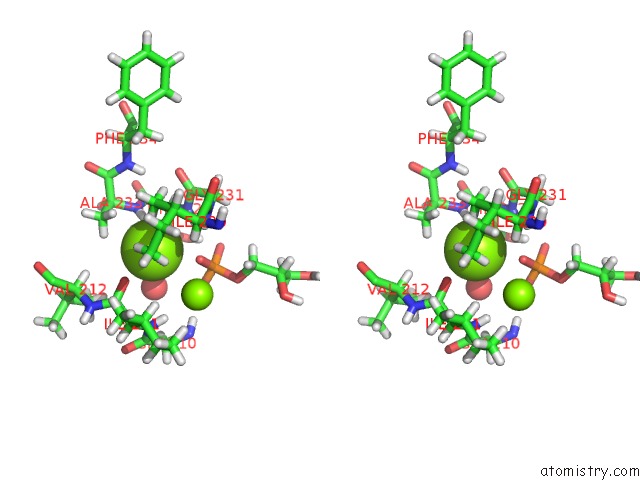
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Gggps From Flavobacterium Johnsoniae within 5.0Å range:
|
Reference:
D.Peterhoff,
B.Beer,
C.Rajendran,
E.P.Kumpula,
E.Kapetaniou,
H.Guldan,
R.K.Wierenga,
R.Sterner,
P.Babinger.
A Comprehensive Analysis of the Geranylgeranylglyceryl Phosphate Synthase Enzyme Family Identifies Novel Members and Reveals Mechanisms of Substrate Specificity and Quaternary Structure Organization. Mol.Microbiol. V. 92 885 2014.
ISSN: ISSN 0950-382X
PubMed: 24684232
DOI: 10.1111/MMI.12596
Page generated: Mon Aug 11 14:41:41 2025
ISSN: ISSN 0950-382X
PubMed: 24684232
DOI: 10.1111/MMI.12596
Last articles
Mg in 4JP1Mg in 4JND
Mg in 4JNE
Mg in 4JKL
Mg in 4JN4
Mg in 4JN9
Mg in 4JLZ
Mg in 4JMG
Mg in 4JKH
Mg in 4JLV