Magnesium »
PDB 4to0-4tyq »
4tqs »
Magnesium in PDB 4tqs: Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp
Enzymatic activity of Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp
All present enzymatic activity of Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp:
2.7.7.7;
2.7.7.7;
Protein crystallography data
The structure of Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp, PDB code: 4tqs
was solved by
L.Zhao,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 28.41 / 2.06 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 52.120, 184.242, 52.124, 90.00, 110.09, 90.00 |
| R / Rfree (%) | 22.8 / 29 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp
(pdb code 4tqs). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 4 binding sites of Magnesium where determined in the Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp, PDB code: 4tqs:
Jump to Magnesium binding site number: 1; 2; 3; 4;
In total 4 binding sites of Magnesium where determined in the Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp, PDB code: 4tqs:
Jump to Magnesium binding site number: 1; 2; 3; 4;
Magnesium binding site 1 out of 4 in 4tqs
Go back to
Magnesium binding site 1 out
of 4 in the Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp
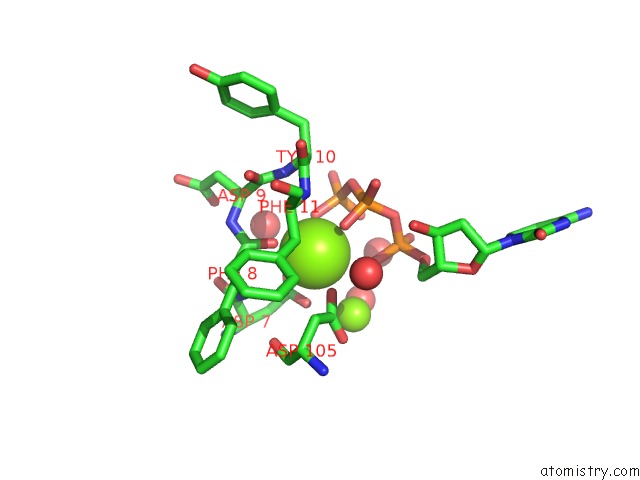
Mono view
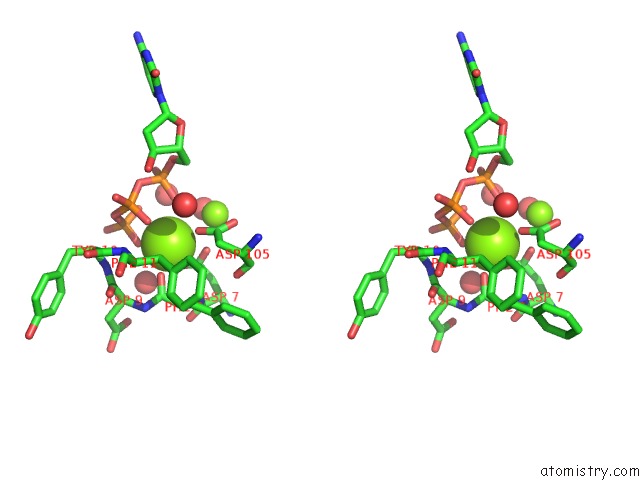
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp within 5.0Å range:
|
Magnesium binding site 2 out of 4 in 4tqs
Go back to
Magnesium binding site 2 out
of 4 in the Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp within 5.0Å range:
|
Magnesium binding site 3 out of 4 in 4tqs
Go back to
Magnesium binding site 3 out
of 4 in the Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp within 5.0Å range:
|
Magnesium binding site 4 out of 4 in 4tqs
Go back to
Magnesium binding site 4 out
of 4 in the Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Ternary Complex of Y-Family Dna Polymerase DPO4 with (5'S)-8,5'-Cyclo- 2'-Deoxyguanosine and Dctp within 5.0Å range:
|
Reference:
W.Xu,
A.M.Ouellette,
Z.Wawrzak,
S.J.Shriver,
S.M.Anderson,
L.Zhao.
Kinetic and Structural Mechanisms of (5'S)-8,5'-Cyclo-2'-Deoxyguanosine-Induced Dna Replication Stalling. Biochemistry V. 54 639 2015.
ISSN: ISSN 0006-2960
PubMed: 25569151
DOI: 10.1021/BI5014936
Page generated: Tue Aug 12 00:11:30 2025
ISSN: ISSN 0006-2960
PubMed: 25569151
DOI: 10.1021/BI5014936
Last articles
Mg in 5ABMMg in 5AC0
Mg in 5ABY
Mg in 5ABN
Mg in 5A9K
Mg in 5AAR
Mg in 5A94
Mg in 5A95
Mg in 5A9F
Mg in 5A99