Magnesium »
PDB 6aad-6ak7 »
6agp »
Magnesium in PDB 6agp: Structure of RAC1 in the Low-Affinity State For MG2+
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of RAC1 in the Low-Affinity State For MG2+
(pdb code 6agp). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of RAC1 in the Low-Affinity State For MG2+, PDB code: 6agp:
In total only one binding site of Magnesium was determined in the Structure of RAC1 in the Low-Affinity State For MG2+, PDB code: 6agp:
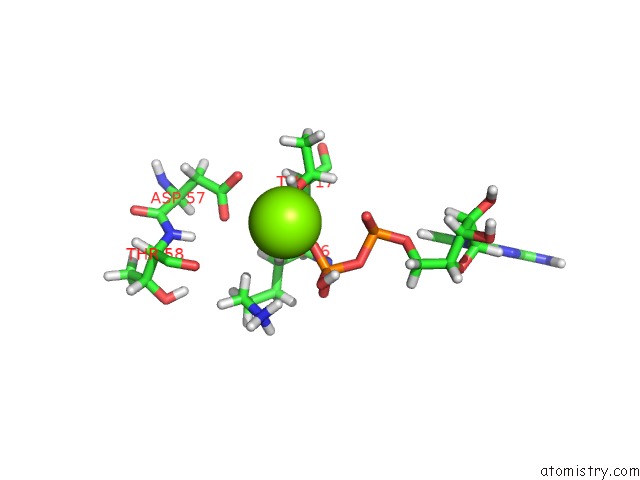
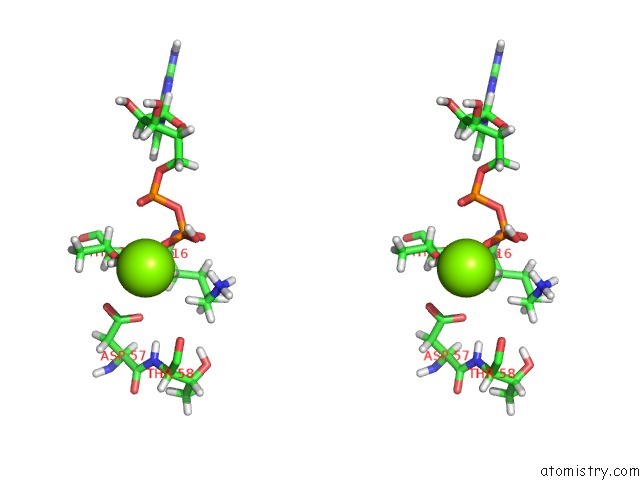
Magnesium binding site 1 out of 1 in 6agp
Go back to
Magnesium binding site 1 out
of 1 in the Structure of RAC1 in the Low-Affinity State For MG2+
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of RAC1 in the Low-Affinity State For MG2+ within 5.0Å range:
|
Reference:
Y.Toyama,
K.Kontani,
T.Katada,
I.Shimada.
Conformational Landscape Alternations Promote Oncogenic Activities of Ras-Related C3 Botulinum Toxin Substrate 1 As Revealed By uc(Nmr). Sci Adv V. 5 V8945 2019.
ISSN: ESSN 2375-2548
PubMed: 30891502
DOI: 10.1126/SCIADV.AAV8945
Page generated: Mon Sep 30 19:12:35 2024
ISSN: ESSN 2375-2548
PubMed: 30891502
DOI: 10.1126/SCIADV.AAV8945
Last articles
Zn in 9JYWZn in 9IR4
Zn in 9IR3
Zn in 9GMX
Zn in 9GMW
Zn in 9JEJ
Zn in 9ERF
Zn in 9ERE
Zn in 9EGV
Zn in 9EGW