Magnesium »
PDB 6iag-6ik9 »
6ii6 »
Magnesium in PDB 6ii6: Crystal Structure of the Makes Caterpillars Floppy (Mcf)-Like Effector of Vibrio Vulnificus MO6-24/O in Complex with A Human Adp- Ribosylation Factor 3 (ARF3)
Protein crystallography data
The structure of Crystal Structure of the Makes Caterpillars Floppy (Mcf)-Like Effector of Vibrio Vulnificus MO6-24/O in Complex with A Human Adp- Ribosylation Factor 3 (ARF3), PDB code: 6ii6
was solved by
Y.Lee,
B.S.Kim,
S.Choi,
E.Y.Lee,
S.Park,
J.Hwang,
Y.Kwon,
J.Hyun,
C.Lee,
S.H.Eom,
M.H.Kim,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.10 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 53.293, 117.523, 186.348, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.5 / 23.2 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Makes Caterpillars Floppy (Mcf)-Like Effector of Vibrio Vulnificus MO6-24/O in Complex with A Human Adp- Ribosylation Factor 3 (ARF3)
(pdb code 6ii6). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Makes Caterpillars Floppy (Mcf)-Like Effector of Vibrio Vulnificus MO6-24/O in Complex with A Human Adp- Ribosylation Factor 3 (ARF3), PDB code: 6ii6:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Makes Caterpillars Floppy (Mcf)-Like Effector of Vibrio Vulnificus MO6-24/O in Complex with A Human Adp- Ribosylation Factor 3 (ARF3), PDB code: 6ii6:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 6ii6
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of the Makes Caterpillars Floppy (Mcf)-Like Effector of Vibrio Vulnificus MO6-24/O in Complex with A Human Adp- Ribosylation Factor 3 (ARF3)
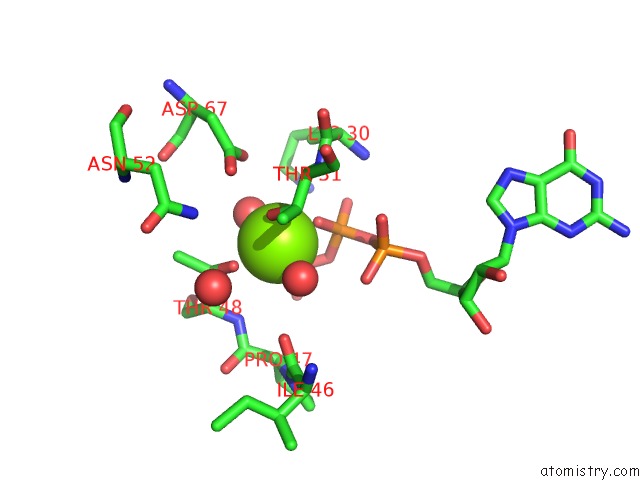
Mono view
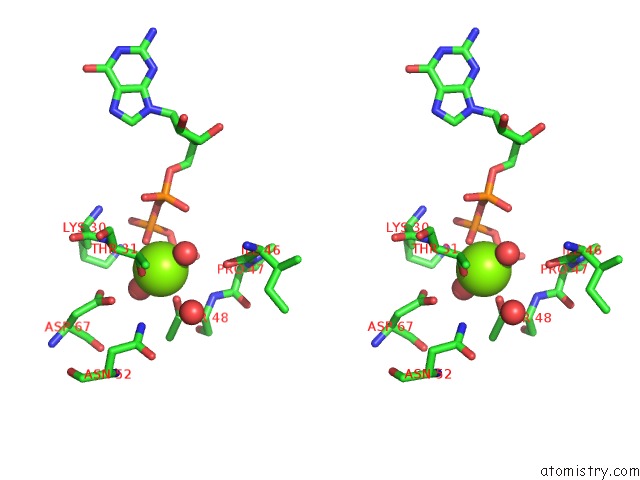
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Makes Caterpillars Floppy (Mcf)-Like Effector of Vibrio Vulnificus MO6-24/O in Complex with A Human Adp- Ribosylation Factor 3 (ARF3) within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 6ii6
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of the Makes Caterpillars Floppy (Mcf)-Like Effector of Vibrio Vulnificus MO6-24/O in Complex with A Human Adp- Ribosylation Factor 3 (ARF3)
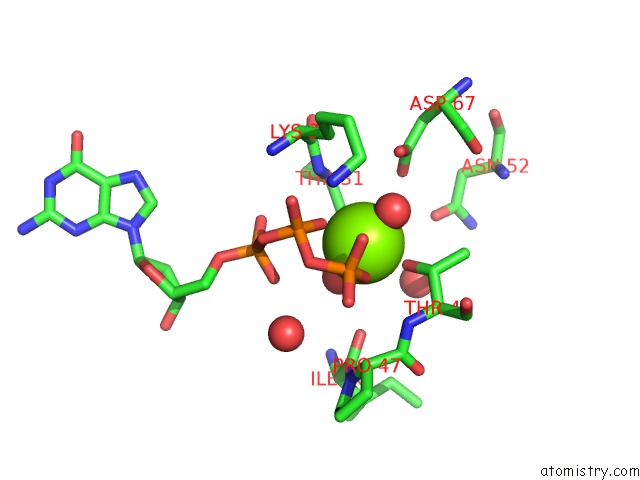
Mono view
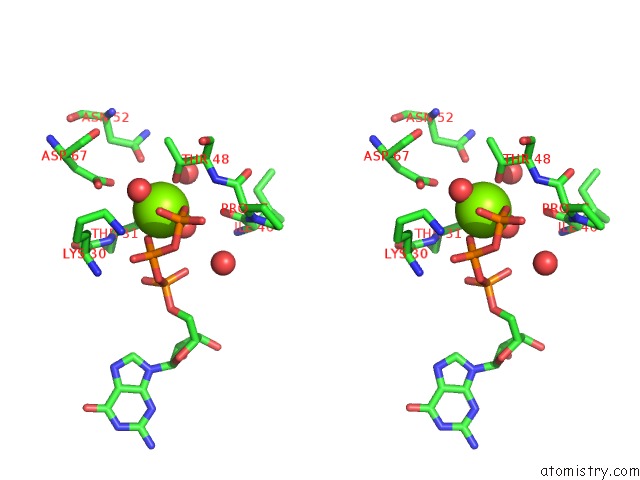
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Makes Caterpillars Floppy (Mcf)-Like Effector of Vibrio Vulnificus MO6-24/O in Complex with A Human Adp- Ribosylation Factor 3 (ARF3) within 5.0Å range:
|
Reference:
Y.Lee,
B.S.Kim,
S.Choi,
E.Y.Lee,
S.Park,
J.Hwang,
Y.Kwon,
J.Hyun,
C.Lee,
S.H.Eom,
M.H.Kim.
Crystal Structure of Pathogenic Bacterial Toxin A with A Host Protein To Be Published.
Page generated: Wed Aug 13 08:08:46 2025
Last articles
Mg in 6S36Mg in 6S3C
Mg in 6S2Z
Mg in 6S2U
Mg in 6S0M
Mg in 6S2T
Mg in 6S2J
Mg in 6S23
Mg in 6S0J
Mg in 6S06