Magnesium »
PDB 6lya-6m6a »
6lzi »
Magnesium in PDB 6lzi: Aquifex Aeolicus Mutl Atpase Domain Complexed with Adp
Protein crystallography data
The structure of Aquifex Aeolicus Mutl Atpase Domain Complexed with Adp, PDB code: 6lzi
was solved by
K.Fukui,
K.Izuhara,
T.Yano,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.73 / 1.69 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 35.017, 42.906, 53.845, 80.98, 71.52, 77.71 |
| R / Rfree (%) | 20.2 / 24.3 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Aquifex Aeolicus Mutl Atpase Domain Complexed with Adp
(pdb code 6lzi). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Aquifex Aeolicus Mutl Atpase Domain Complexed with Adp, PDB code: 6lzi:
In total only one binding site of Magnesium was determined in the Aquifex Aeolicus Mutl Atpase Domain Complexed with Adp, PDB code: 6lzi:
Magnesium binding site 1 out of 1 in 6lzi
Go back to
Magnesium binding site 1 out
of 1 in the Aquifex Aeolicus Mutl Atpase Domain Complexed with Adp
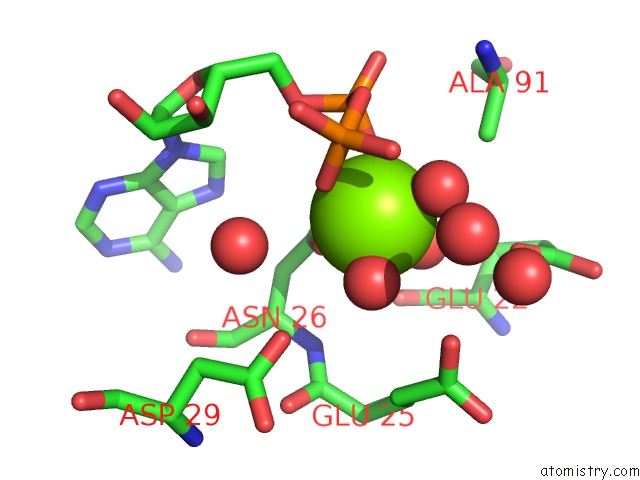
Mono view
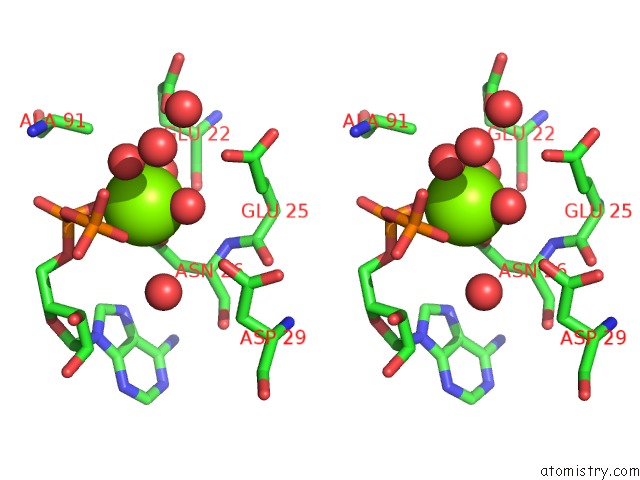
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Aquifex Aeolicus Mutl Atpase Domain Complexed with Adp within 5.0Å range:
|
Reference:
K.Izuhara,
K.Fukui,
T.Murakawa,
S.Baba,
T.Kumasaka,
K.Uchiyama,
T.Yano.
A Lynch Syndrome-Associated Mutation at A Bergerat Atp-Binding Fold Destabilizes the Structure of the Dna Mismatch Repair Endonuclease Mutl. J.Biol.Chem. V. 295 11643 2020.
ISSN: ESSN 1083-351X
PubMed: 32571878
DOI: 10.1074/JBC.RA120.013576
Page generated: Wed Aug 13 11:57:04 2025
ISSN: ESSN 1083-351X
PubMed: 32571878
DOI: 10.1074/JBC.RA120.013576
Last articles
Mg in 7JIGMg in 7JHO
Mg in 7JIF
Mg in 7JHP
Mg in 7JHL
Mg in 7JH7
Mg in 7JHI
Mg in 7JHN
Mg in 7JGS
Mg in 7JH9