Magnesium »
PDB 6lya-6m6a »
6m3y »
Magnesium in PDB 6m3y: Crystal Structure of Pilus Adhesin, Spac From Lactobacillus Rhamnosus Gg - Open Conformation
Protein crystallography data
The structure of Crystal Structure of Pilus Adhesin, Spac From Lactobacillus Rhamnosus Gg - Open Conformation, PDB code: 6m3y
was solved by
A.Kant,
A.Palva,
I.Von Ossowski,
V.Krishnan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 83.53 / 1.93 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.045, 110.023, 128.344, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.7 / 24.8 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Pilus Adhesin, Spac From Lactobacillus Rhamnosus Gg - Open Conformation
(pdb code 6m3y). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Pilus Adhesin, Spac From Lactobacillus Rhamnosus Gg - Open Conformation, PDB code: 6m3y:
In total only one binding site of Magnesium was determined in the Crystal Structure of Pilus Adhesin, Spac From Lactobacillus Rhamnosus Gg - Open Conformation, PDB code: 6m3y:
Magnesium binding site 1 out of 1 in 6m3y
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Pilus Adhesin, Spac From Lactobacillus Rhamnosus Gg - Open Conformation
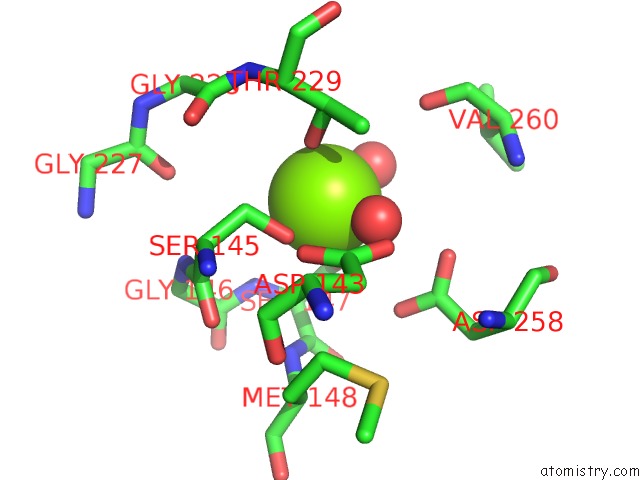
Mono view
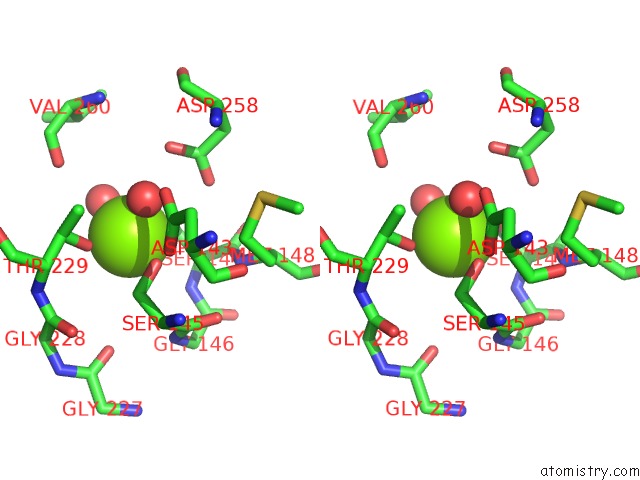
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Pilus Adhesin, Spac From Lactobacillus Rhamnosus Gg - Open Conformation within 5.0Å range:
|
Reference:
A.Kant,
A.Palva,
I.Von Ossowski,
V.Krishnan.
Crystal Structure of Lactobacillar Spac Reveals An Atypical Five-Domain Pilus Tip Adhesin: Exposing Its Substrate-Binding and Assembly in Spacba Pili. J.Struct.Biol. V. 211 07571 2020.
ISSN: ESSN 1095-8657
PubMed: 32653644
DOI: 10.1016/J.JSB.2020.107571
Page generated: Wed Aug 13 11:58:59 2025
ISSN: ESSN 1095-8657
PubMed: 32653644
DOI: 10.1016/J.JSB.2020.107571
Last articles
Mg in 7DUJMg in 7DUI
Mg in 7DUH
Mg in 7DUG
Mg in 7DR0
Mg in 7DR1
Mg in 7DU2
Mg in 7DSP
Mg in 7DSJ
Mg in 7DSI