Magnesium »
PDB 8f8r-8fix »
8fix »
Magnesium in PDB 8fix: Cryo-Em Structure of E. Coli Rna Polymerase Backtracked Elongation Complex Harboring A Terminal Mismatch
Enzymatic activity of Cryo-Em Structure of E. Coli Rna Polymerase Backtracked Elongation Complex Harboring A Terminal Mismatch
All present enzymatic activity of Cryo-Em Structure of E. Coli Rna Polymerase Backtracked Elongation Complex Harboring A Terminal Mismatch:
2.7.7.6;
2.7.7.6;
Other elements in 8fix:
The structure of Cryo-Em Structure of E. Coli Rna Polymerase Backtracked Elongation Complex Harboring A Terminal Mismatch also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of E. Coli Rna Polymerase Backtracked Elongation Complex Harboring A Terminal Mismatch
(pdb code 8fix). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of E. Coli Rna Polymerase Backtracked Elongation Complex Harboring A Terminal Mismatch, PDB code: 8fix:
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of E. Coli Rna Polymerase Backtracked Elongation Complex Harboring A Terminal Mismatch, PDB code: 8fix:
Magnesium binding site 1 out of 1 in 8fix
Go back to
Magnesium binding site 1 out
of 1 in the Cryo-Em Structure of E. Coli Rna Polymerase Backtracked Elongation Complex Harboring A Terminal Mismatch
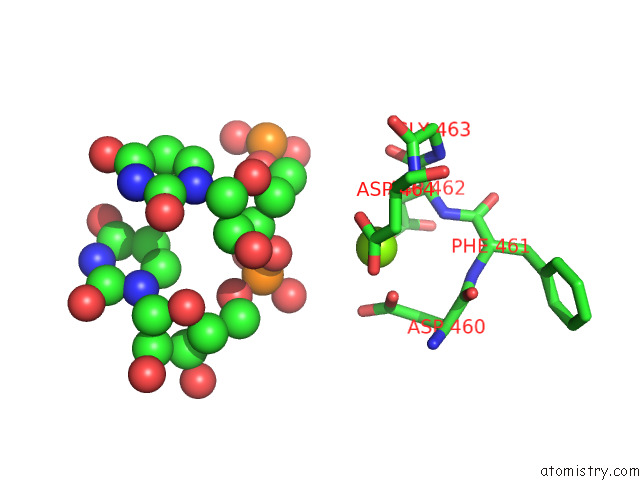
Mono view
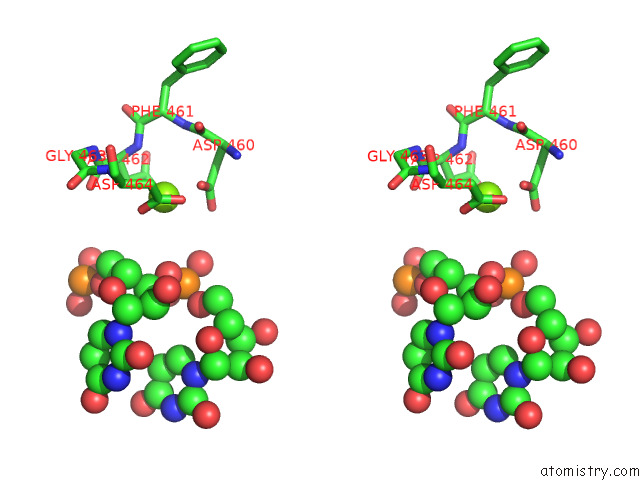
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of E. Coli Rna Polymerase Backtracked Elongation Complex Harboring A Terminal Mismatch within 5.0Å range:
|
Reference:
L.M.Wee,
A.B.Tong,
A.J.Florez Ariza,
C.Canari-Chumpitaz,
P.Grob,
E.Nogales,
C.J.Bustamante.
A Trailing Ribosome Speeds Up Rna Polymerase at the Expense of Transcript Fidelity Via Force and Allostery. Cell V. 186 1244 2023.
ISSN: ISSN 1097-4172
PubMed: 36931247
DOI: 10.1016/J.CELL.2023.02.008
Page generated: Fri Aug 15 04:51:04 2025
ISSN: ISSN 1097-4172
PubMed: 36931247
DOI: 10.1016/J.CELL.2023.02.008
Last articles
Mg in 8HD7Mg in 8HD6
Mg in 8HG1
Mg in 8HF5
Mg in 8HFH
Mg in 8HE6
Mg in 8HE5
Mg in 8HCD
Mg in 8HCC
Mg in 8HCH