Magnesium »
PDB 8osf-8p0g »
8ox5 »
Magnesium in PDB 8ox5: Cryo-Em Structure of ATP8B1-CDC50A in E1P-Adp Conformation
Enzymatic activity of Cryo-Em Structure of ATP8B1-CDC50A in E1P-Adp Conformation
All present enzymatic activity of Cryo-Em Structure of ATP8B1-CDC50A in E1P-Adp Conformation:
7.6.2.1;
7.6.2.1;
Other elements in 8ox5:
The structure of Cryo-Em Structure of ATP8B1-CDC50A in E1P-Adp Conformation also contains other interesting chemical elements:
| Fluorine | (F) | 4 atoms |
| Aluminium | (Al) | 1 atom |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of ATP8B1-CDC50A in E1P-Adp Conformation
(pdb code 8ox5). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of ATP8B1-CDC50A in E1P-Adp Conformation, PDB code: 8ox5:
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of ATP8B1-CDC50A in E1P-Adp Conformation, PDB code: 8ox5:
Magnesium binding site 1 out of 1 in 8ox5
Go back to
Magnesium binding site 1 out
of 1 in the Cryo-Em Structure of ATP8B1-CDC50A in E1P-Adp Conformation
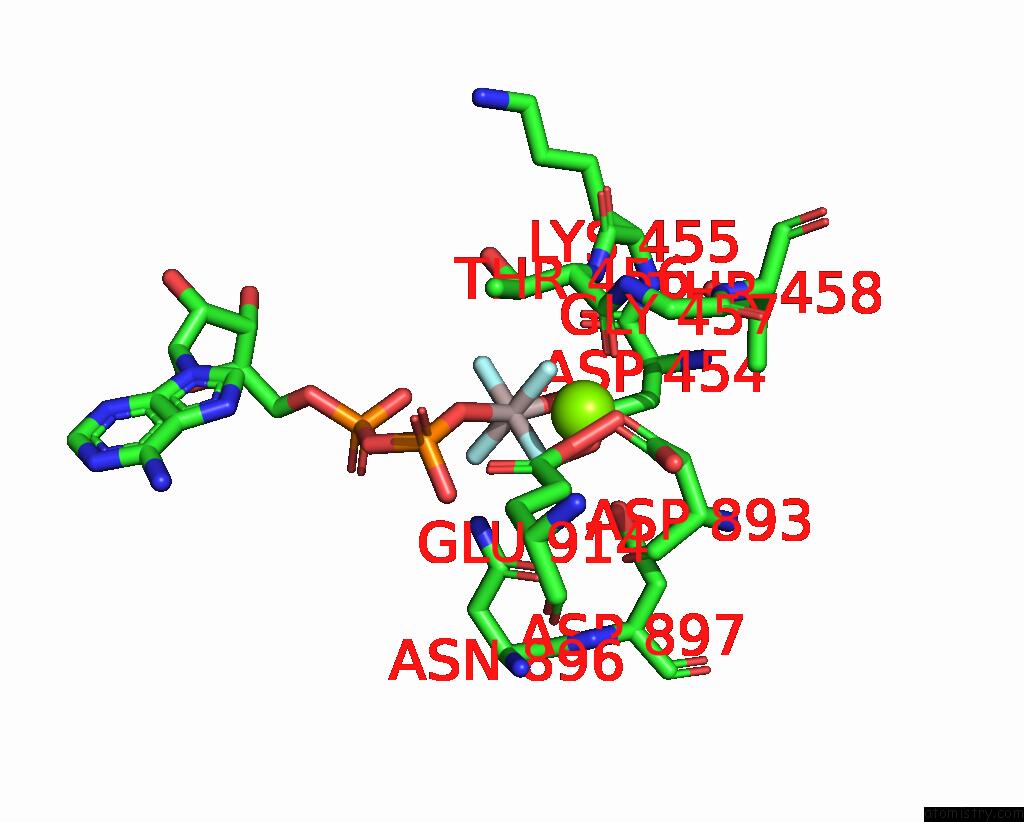
Mono view
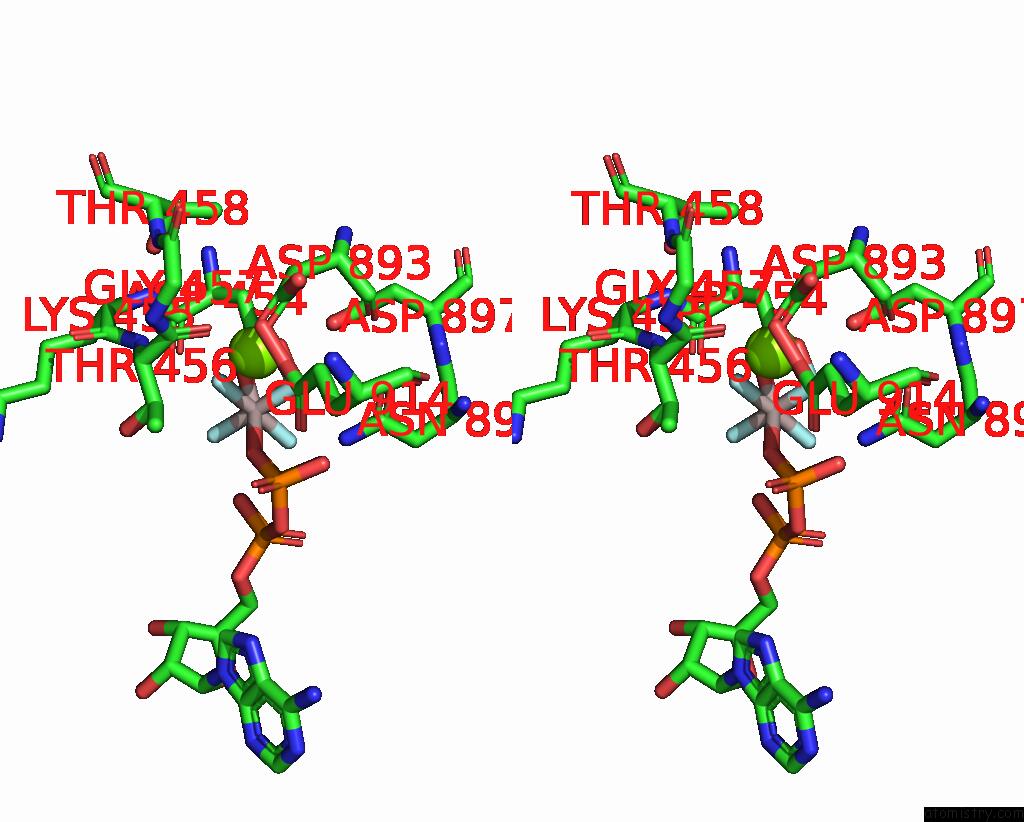
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of ATP8B1-CDC50A in E1P-Adp Conformation within 5.0Å range:
|
Reference:
T.Dieudonne,
F.Kummerer,
M.J.Laursen,
C.Stock,
R.K.Flygaard,
S.Khalid,
G.Lenoir,
J.A.Lyons,
K.Lindorff-Larsen,
P.Nissen.
Activation and Substrate Specificity of the Human P4-Atpase ATP8B1. Nat Commun V. 14 7492 2023.
ISSN: ESSN 2041-1723
PubMed: 37980352
DOI: 10.1038/S41467-023-42828-9
Page generated: Fri Aug 15 11:42:10 2025
ISSN: ESSN 2041-1723
PubMed: 37980352
DOI: 10.1038/S41467-023-42828-9
Last articles
Mg in 8RJ6Mg in 8RHL
Mg in 8RJ4
Mg in 8RIZ
Mg in 8RIX
Mg in 8RIK
Mg in 8RDU
Mg in 8RI5
Mg in 8RI1
Mg in 8RI3