Magnesium »
PDB 1eo3-1f6t »
1f6t »
Magnesium in PDB 1f6t: Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex
Enzymatic activity of Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex
All present enzymatic activity of Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex:
2.7.4.6;
2.7.4.6;
Protein crystallography data
The structure of Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex, PDB code: 1f6t
was solved by
C.Guerreiro,
J.Boretto,
J.Janin,
M.Veron,
B.Canard,
B.Schneider,
S.Sarfati,
D.Deville-Bonne,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.92 |
| Space group | P 31 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 70.359, 70.359, 153.792, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 22.8 / 27.4 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex
(pdb code 1f6t). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex, PDB code: 1f6t:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex, PDB code: 1f6t:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 1f6t
Go back to
Magnesium binding site 1 out
of 3 in the Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex
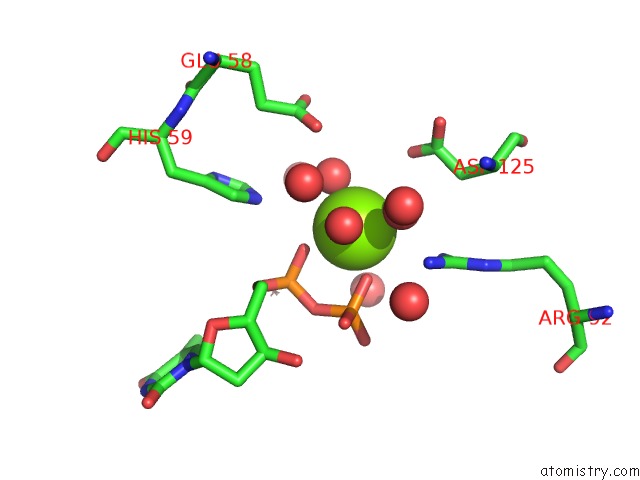
Mono view
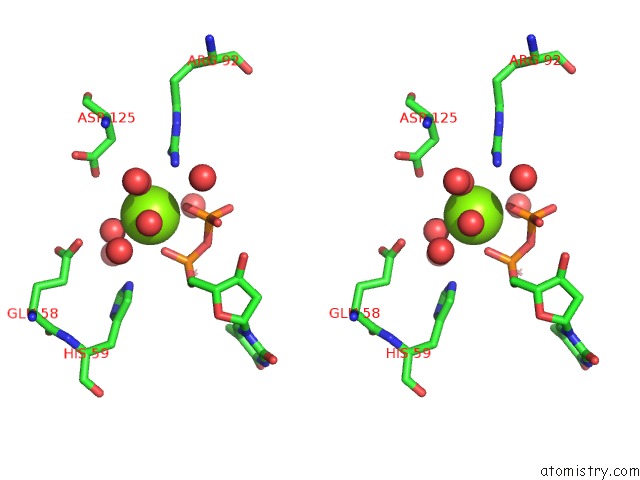
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 1f6t
Go back to
Magnesium binding site 2 out
of 3 in the Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex
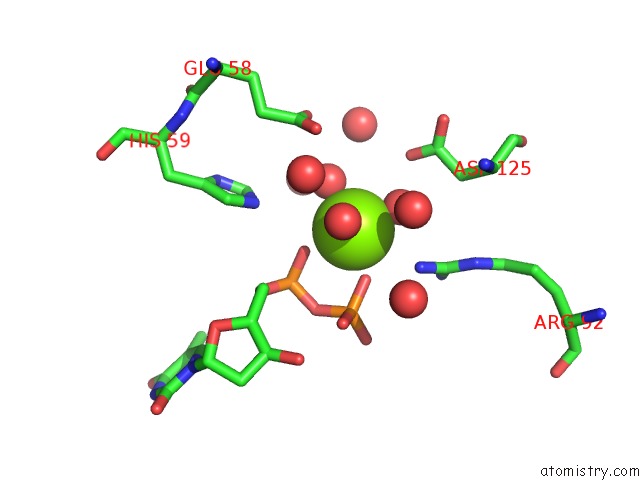
Mono view
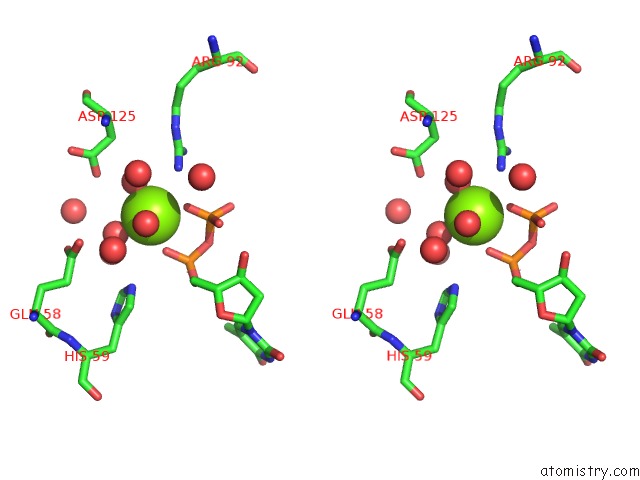
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 1f6t
Go back to
Magnesium binding site 3 out
of 3 in the Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex
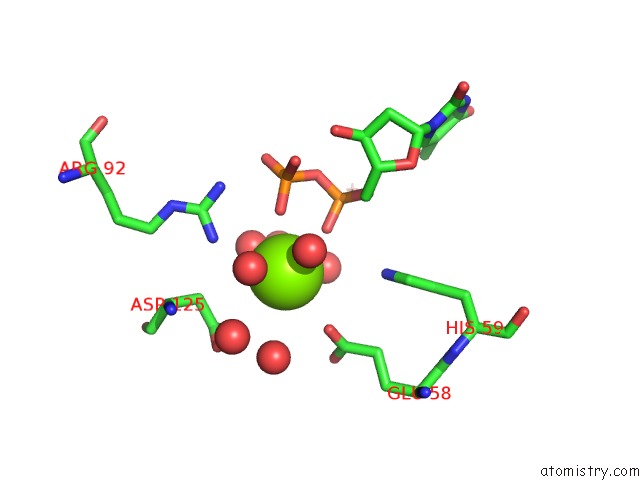
Mono view
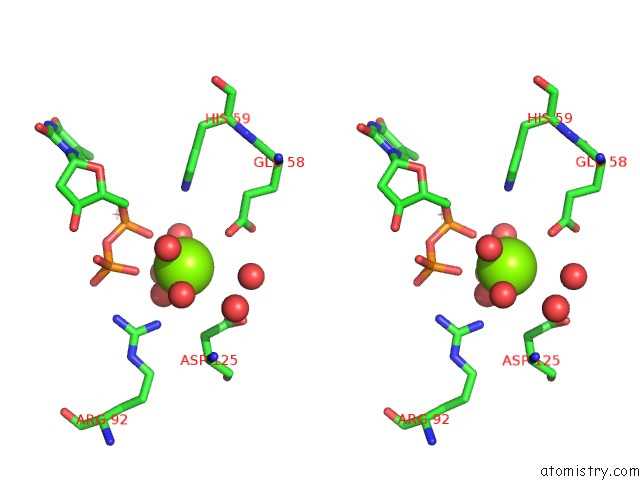
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Structure of the Nucleoside Diphosphate Kinase/Alpha-Borano(Rp)-Tdp.Mg Complex within 5.0Å range:
|
Reference:
P.Meyer,
B.Schneider,
S.Sarfati,
D.Deville-Bonne,
C.Guerreiro,
J.Boretto,
J.Janin,
M.Veron,
B.Canard.
Structural Basis For Activation of Alpha-Boranophosphate Nucleotide Analogues Targeting Drug-Resistant Reverse Transcriptase. Embo J. V. 19 3520 2000.
ISSN: ISSN 0261-4189
PubMed: 10899107
DOI: 10.1093/EMBOJ/19.14.3520
Page generated: Sat Aug 9 20:54:07 2025
ISSN: ISSN 0261-4189
PubMed: 10899107
DOI: 10.1093/EMBOJ/19.14.3520
Last articles
Mg in 4PL3Mg in 4PL0
Mg in 4PKD
Mg in 4PJR
Mg in 4PK4
Mg in 4PJN
Mg in 4PJL
Mg in 4PJK
Mg in 4PJM
Mg in 4PJ0