Magnesium »
PDB 6akp-6asw »
6ap4 »
Magnesium in PDB 6ap4: Crystal Structure of the Dna Polymerase III Subunit Beta From Acinetobacter Baumannii
Protein crystallography data
The structure of Crystal Structure of the Dna Polymerase III Subunit Beta From Acinetobacter Baumannii, PDB code: 6ap4
was solved by
A.E.Mcgrath,
A.J.Oakley,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 164.30 / 2.95 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 72.713, 328.597, 147.497, 90.00, 91.53, 90.00 |
| R / Rfree (%) | 24.8 / 28.7 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Dna Polymerase III Subunit Beta From Acinetobacter Baumannii
(pdb code 6ap4). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Dna Polymerase III Subunit Beta From Acinetobacter Baumannii, PDB code: 6ap4:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of the Dna Polymerase III Subunit Beta From Acinetobacter Baumannii, PDB code: 6ap4:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 6ap4
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of the Dna Polymerase III Subunit Beta From Acinetobacter Baumannii
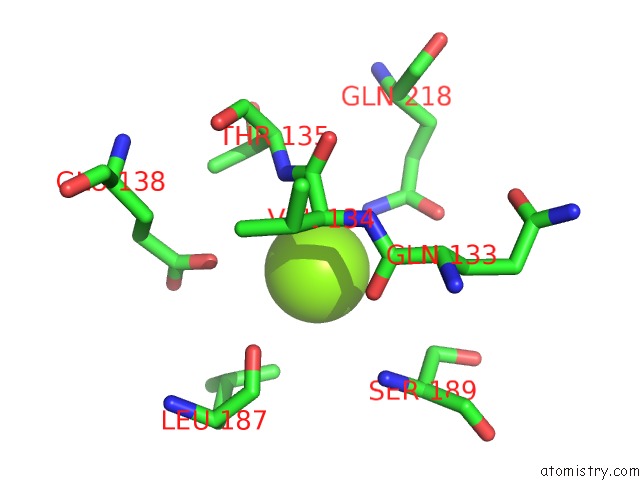
Mono view
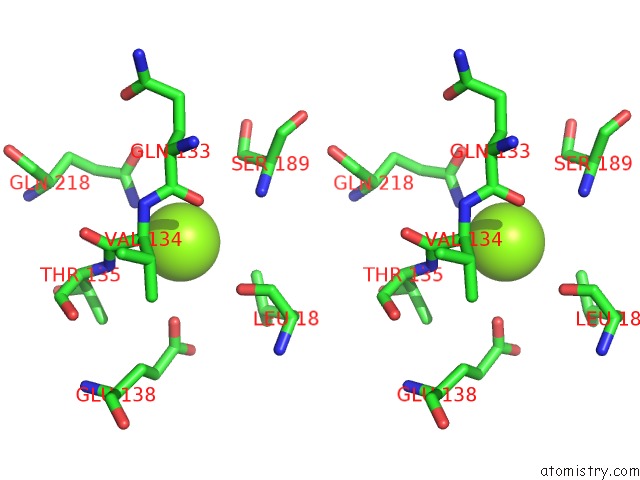
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Dna Polymerase III Subunit Beta From Acinetobacter Baumannii within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 6ap4
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of the Dna Polymerase III Subunit Beta From Acinetobacter Baumannii
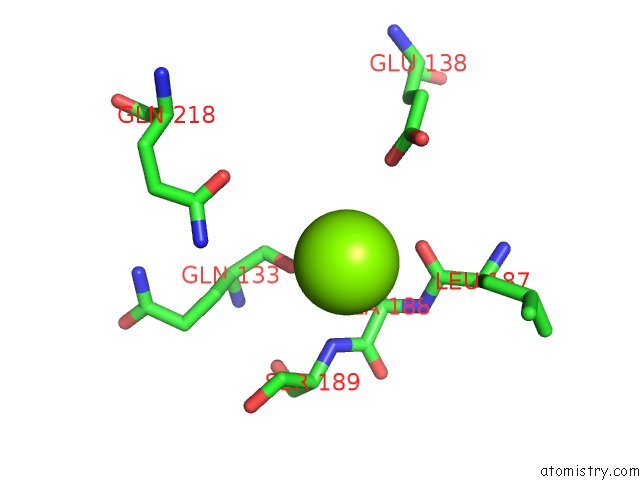
Mono view
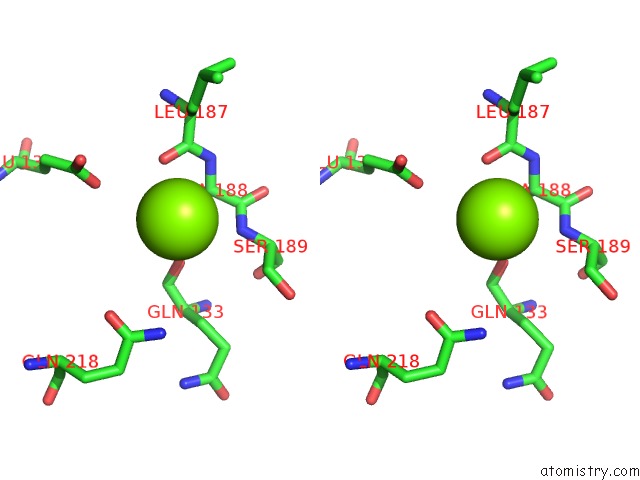
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Dna Polymerase III Subunit Beta From Acinetobacter Baumannii within 5.0Å range:
|
Reference:
A.E.Mcgrath,
A.P.Martyn,
L.R.Whittell,
F.E.Dawes,
J.L.Beck,
N.E.Dixon,
M.J.Kelso,
A.J.Oakley.
Crystal Structures and Biochemical Characterization of Dna Sliding Clamps From Three Gram-Negative Bacterial Pathogens. J. Struct. Biol. V. 204 396 2018.
ISSN: ESSN 1095-8657
PubMed: 30366028
DOI: 10.1016/J.JSB.2018.10.008
Page generated: Mon Sep 30 19:16:19 2024
ISSN: ESSN 1095-8657
PubMed: 30366028
DOI: 10.1016/J.JSB.2018.10.008
Last articles
Mg in 5GWJMg in 5GWI
Mg in 5GW7
Mg in 5GVE
Mg in 5GVX
Mg in 5GVD
Mg in 5GUL
Mg in 5GVC
Mg in 5GV2
Mg in 5GUH