Magnesium »
PDB 6dwd-6e9e »
6e47 »
Magnesium in PDB 6e47: Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid
Protein crystallography data
The structure of Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid, PDB code: 6e47
was solved by
C.A.Nelson,
D.H.Fremont,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 60.70 / 1.95 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 49.105, 134.824, 139.592, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.8 / 25.7 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid
(pdb code 6e47). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 6 binding sites of Magnesium where determined in the Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid, PDB code: 6e47:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Magnesium where determined in the Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid, PDB code: 6e47:
Jump to Magnesium binding site number: 1; 2; 3; 4; 5; 6;
Magnesium binding site 1 out of 6 in 6e47
Go back to
Magnesium binding site 1 out
of 6 in the Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid within 5.0Å range:
|
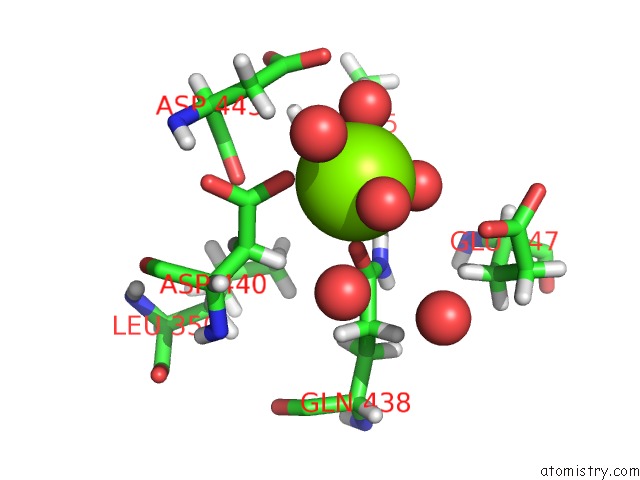
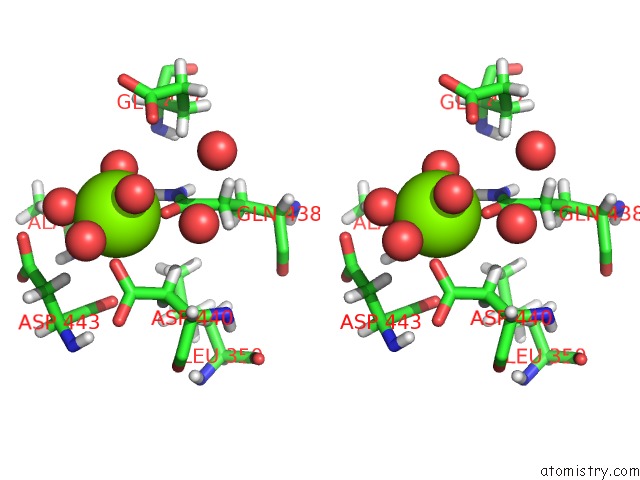
Magnesium binding site 2 out of 6 in 6e47
Go back to
Magnesium binding site 2 out
of 6 in the Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid within 5.0Å range:
|
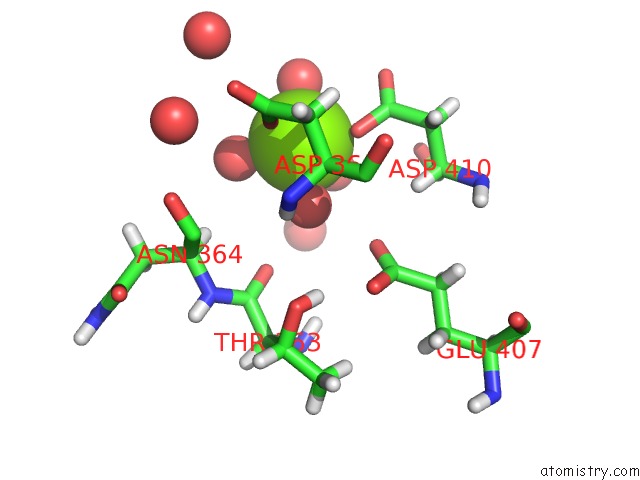
Magnesium binding site 3 out of 6 in 6e47
Go back to
Magnesium binding site 3 out
of 6 in the Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid within 5.0Å range:
|
Magnesium binding site 4 out of 6 in 6e47
Go back to
Magnesium binding site 4 out
of 6 in the Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 4 of Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid within 5.0Å range:
|
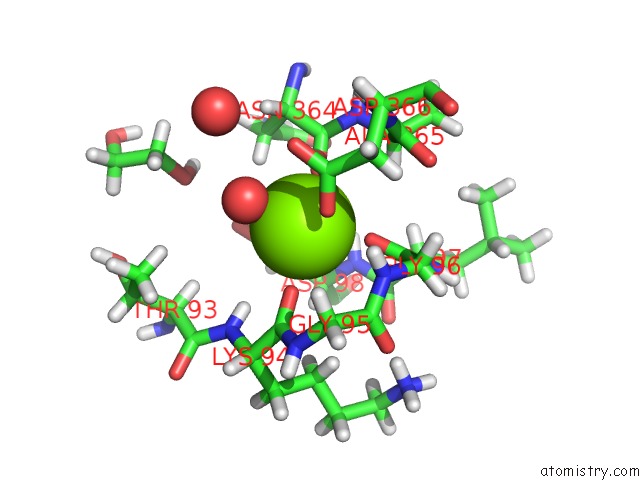
Magnesium binding site 5 out of 6 in 6e47
Go back to
Magnesium binding site 5 out
of 6 in the Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 5 of Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid within 5.0Å range:
|
Magnesium binding site 6 out of 6 in 6e47
Go back to
Magnesium binding site 6 out
of 6 in the Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 6 of Crystal Structure of the Murine Norovirus VP1 P Domain in Complex with the CD300LF Receptor and Glycochenodeoxycholic Acid within 5.0Å range:
|
Reference:
C.A.Nelson,
C.B.Wilen,
Y.N.Dai,
R.C.Orchard,
A.S.Kim,
R.A.Stegeman,
L.L.Hsieh,
T.J.Smith,
H.W.Virgin,
D.H.Fremont.
Structural Basis For Murine Norovirus Engagement of Bile Acids and the CD300LF Receptor. Proc. Natl. Acad. Sci. V. 115 E9201 2018U.S.A..
ISSN: ESSN 1091-6490
PubMed: 30194229
DOI: 10.1073/PNAS.1805797115
Page generated: Wed Aug 13 05:36:34 2025
ISSN: ESSN 1091-6490
PubMed: 30194229
DOI: 10.1073/PNAS.1805797115
Last articles
F in 9RF5F in 9N6R
F in 9N6P
F in 9N6M
F in 9GSG
F in 9GSD
F in 9GSY
F in 9D02
F in 9D03
F in 9D04