Magnesium »
PDB 1gq9-1h7q »
1gsa »
Magnesium in PDB 1gsa: Structure of Glutathione Synthetase Complexed with Adp and Glutathione
Enzymatic activity of Structure of Glutathione Synthetase Complexed with Adp and Glutathione
All present enzymatic activity of Structure of Glutathione Synthetase Complexed with Adp and Glutathione:
6.3.2.3;
6.3.2.3;
Protein crystallography data
The structure of Structure of Glutathione Synthetase Complexed with Adp and Glutathione, PDB code: 1gsa
was solved by
T.Hara,
H.Kato,
T.Nishioka,
Y.Katsube,
J.Oda,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 8.00 / 2.00 |
| Space group | P 62 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 87.250, 87.250, 169.580, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 18.8 / n/a |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of Glutathione Synthetase Complexed with Adp and Glutathione
(pdb code 1gsa). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Structure of Glutathione Synthetase Complexed with Adp and Glutathione, PDB code: 1gsa:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Structure of Glutathione Synthetase Complexed with Adp and Glutathione, PDB code: 1gsa:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 1gsa
Go back to
Magnesium binding site 1 out
of 2 in the Structure of Glutathione Synthetase Complexed with Adp and Glutathione
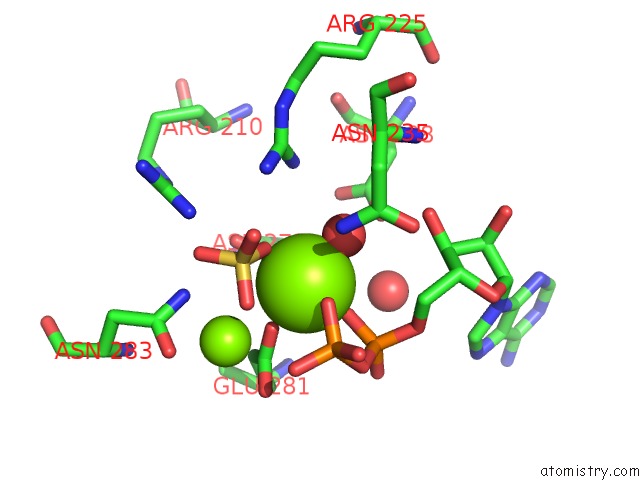
Mono view
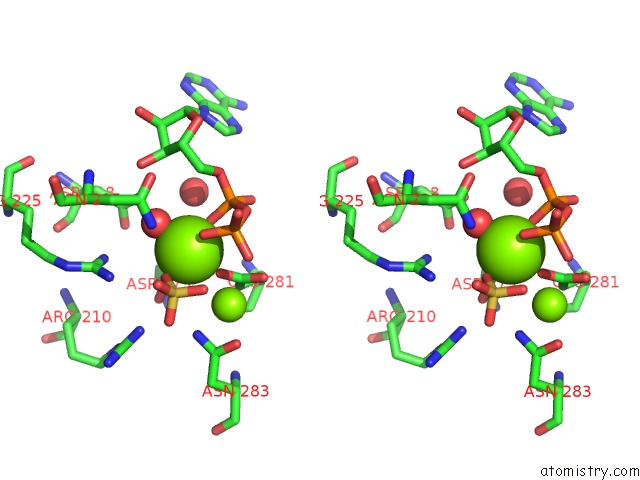
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of Glutathione Synthetase Complexed with Adp and Glutathione within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 1gsa
Go back to
Magnesium binding site 2 out
of 2 in the Structure of Glutathione Synthetase Complexed with Adp and Glutathione
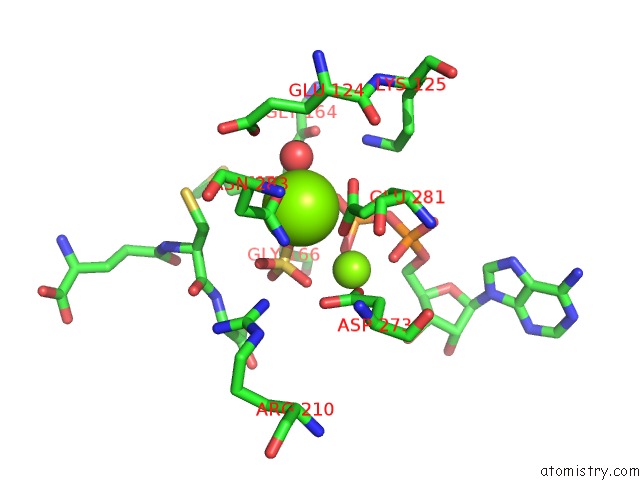
Mono view
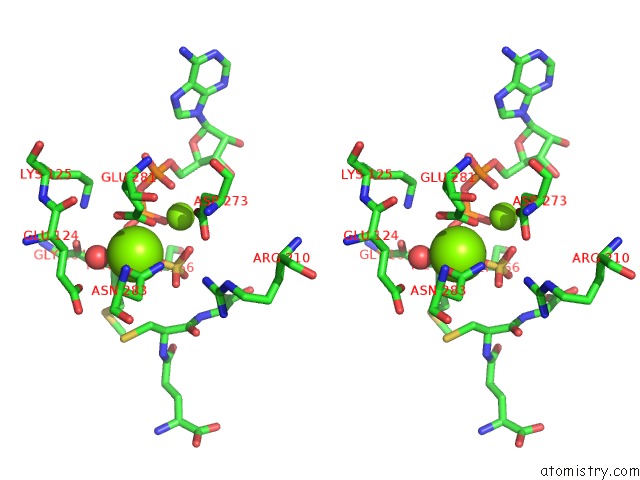
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Structure of Glutathione Synthetase Complexed with Adp and Glutathione within 5.0Å range:
|
Reference:
T.Hara,
H.Kato,
Y.Katsube,
J.Oda.
A Pseudo-Michaelis Quaternary Complex in the Reverse Reaction of A Ligase: Structure of Escherichia Coli B Glutathione Synthetase Complexed with Adp, Glutathione, and Sulfate at 2.0 A Resolution. Biochemistry V. 35 11967 1996.
ISSN: ISSN 0006-2960
PubMed: 8810901
DOI: 10.1021/BI9605245
Page generated: Sat Aug 9 21:23:04 2025
ISSN: ISSN 0006-2960
PubMed: 8810901
DOI: 10.1021/BI9605245
Last articles
Mg in 3JAYMg in 3JB3
Mg in 3JB2
Mg in 3JAP
Mg in 3JAM
Mg in 3JAT
Mg in 3JAW
Mg in 3JAS
Mg in 3JAR
Mg in 3J81