Magnesium »
PDB 5bjp-5btm »
5bmp »
Magnesium in PDB 5bmp: Crystal Structure of Phosphoglucomutase From Xanthomonas Citri Complexed with Glucose-1-Phosphate
Enzymatic activity of Crystal Structure of Phosphoglucomutase From Xanthomonas Citri Complexed with Glucose-1-Phosphate
All present enzymatic activity of Crystal Structure of Phosphoglucomutase From Xanthomonas Citri Complexed with Glucose-1-Phosphate:
5.4.2.2;
5.4.2.2;
Protein crystallography data
The structure of Crystal Structure of Phosphoglucomutase From Xanthomonas Citri Complexed with Glucose-1-Phosphate, PDB code: 5bmp
was solved by
L.S.Goto,
H.M.Pereira,
M.T.M.Novo Mansur,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.56 / 1.85 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.861, 54.732, 173.143, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.2 / 18 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Phosphoglucomutase From Xanthomonas Citri Complexed with Glucose-1-Phosphate
(pdb code 5bmp). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Crystal Structure of Phosphoglucomutase From Xanthomonas Citri Complexed with Glucose-1-Phosphate, PDB code: 5bmp:
In total only one binding site of Magnesium was determined in the Crystal Structure of Phosphoglucomutase From Xanthomonas Citri Complexed with Glucose-1-Phosphate, PDB code: 5bmp:
Magnesium binding site 1 out of 1 in 5bmp
Go back to
Magnesium binding site 1 out
of 1 in the Crystal Structure of Phosphoglucomutase From Xanthomonas Citri Complexed with Glucose-1-Phosphate
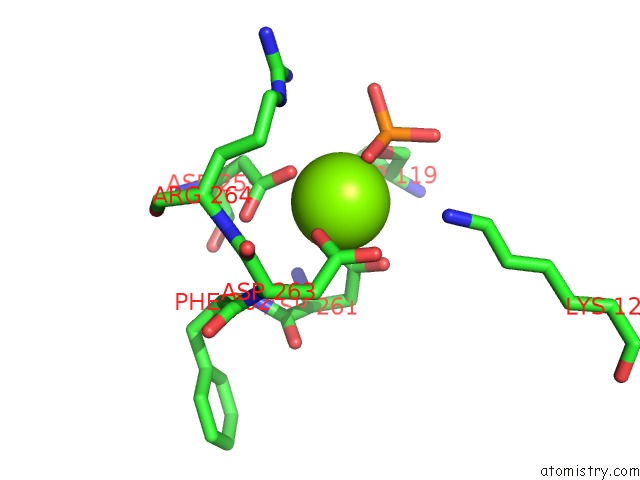
Mono view
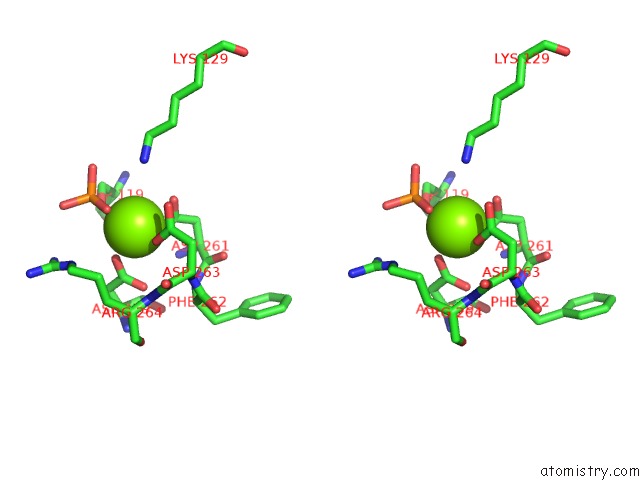
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Phosphoglucomutase From Xanthomonas Citri Complexed with Glucose-1-Phosphate within 5.0Å range:
|
Reference:
L.S.Goto,
A.Vessoni Alexandrino,
C.Malvessi Pereira,
C.Silva Martins,
H.D'muniz Pereira,
J.Brandao-Neto,
M.T.Marques Novo-Mansur.
Structural and Functional Characterization of the Phosphoglucomutase From Xanthomonas Citri Subsp. Citri. Biochim.Biophys.Acta V.1864 1658 2016.
ISSN: ISSN 0006-3002
PubMed: 27567706
DOI: 10.1016/J.BBAPAP.2016.08.014
Page generated: Tue Aug 12 05:41:33 2025
ISSN: ISSN 0006-3002
PubMed: 27567706
DOI: 10.1016/J.BBAPAP.2016.08.014
Last articles
Mg in 5HPOMg in 5HPH
Mg in 5HOO
Mg in 5HOK
Mg in 5HOB
Mg in 5HO5
Mg in 5HMQ
Mg in 5HNZ
Mg in 5HNY
Mg in 5HNX