Magnesium »
PDB 6j7r-6jlj »
6jdk »
Magnesium in PDB 6jdk: Crystal Structure of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans
Protein crystallography data
The structure of Crystal Structure of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans, PDB code: 6jdk
was solved by
J.-S.Kim,
T.D.Nguyen,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.88 / 2.50 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 90.831, 55.049, 118.099, 90.00, 95.25, 90.00 |
| R / Rfree (%) | 20.7 / 27 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Crystal Structure of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans
(pdb code 6jdk). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans, PDB code: 6jdk:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Crystal Structure of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans, PDB code: 6jdk:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 6jdk
Go back to
Magnesium binding site 1 out
of 2 in the Crystal Structure of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans
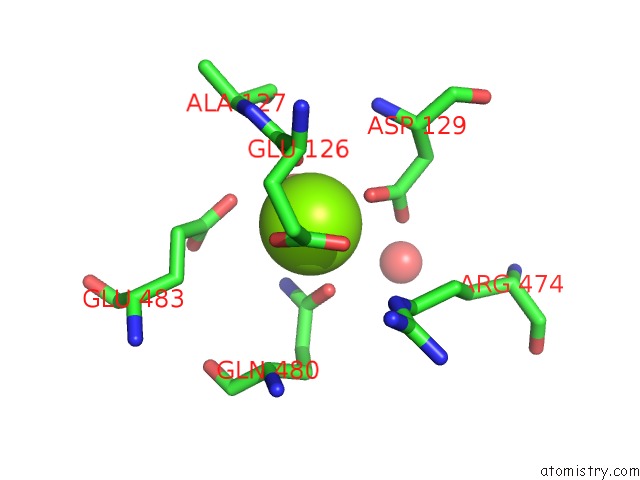
Mono view
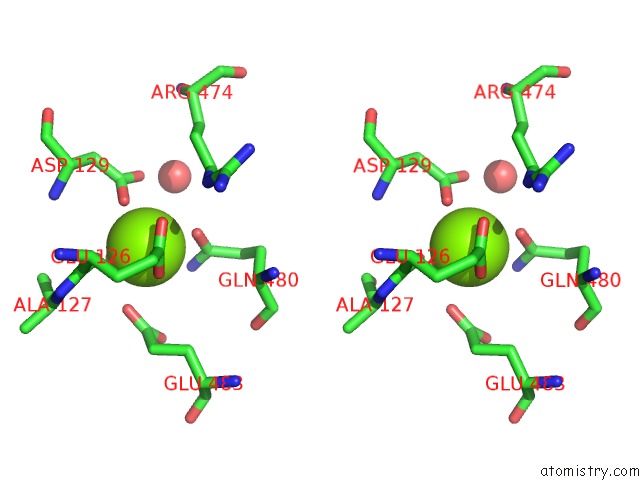
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Crystal Structure of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 6jdk
Go back to
Magnesium binding site 2 out
of 2 in the Crystal Structure of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans
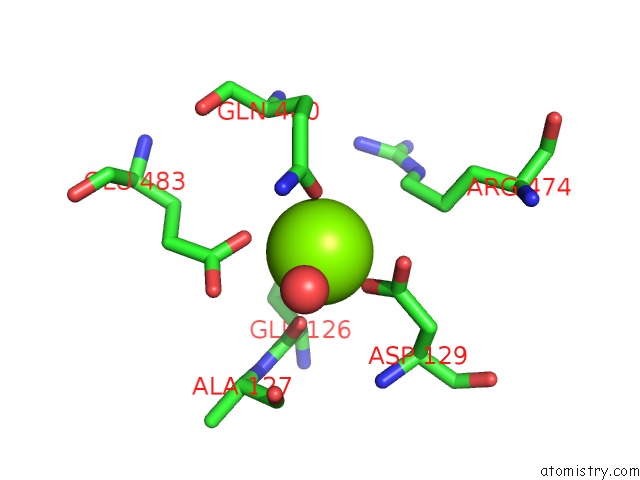
Mono view
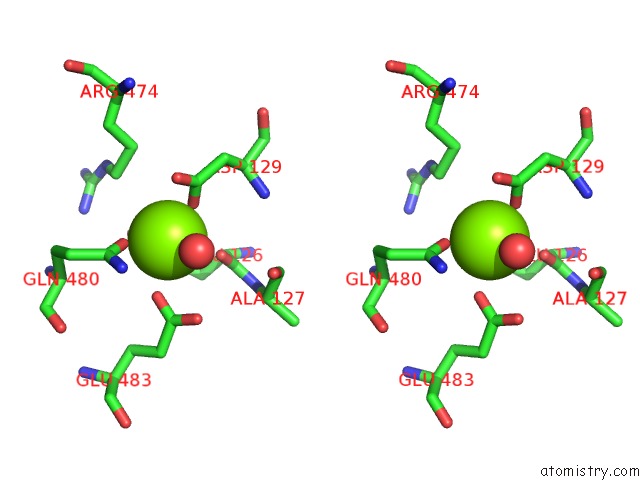
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Crystal Structure of Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans within 5.0Å range:
|
Reference:
T.D.Nguyen,
G.E.Choi,
D.H.Gu,
P.W.Seo,
J.W.Kim,
J.B.Park,
J.S.Kim.
Structural Basis For the Selective Addition of An Oxygen Atom to Cyclic Ketones By Baeyer-Villiger Monooxygenase From Parvibaculum Lavamentivorans. Biochem. Biophys. Res. V. 512 564 2019COMMUN..
ISSN: ESSN 1090-2104
PubMed: 30914200
DOI: 10.1016/J.BBRC.2019.03.114
Page generated: Wed Aug 13 09:11:27 2025
ISSN: ESSN 1090-2104
PubMed: 30914200
DOI: 10.1016/J.BBRC.2019.03.114
Last articles
Mg in 7A17Mg in 7A0V
Mg in 6ZZX
Mg in 6ZXS
Mg in 7A0Q
Mg in 7A0P
Mg in 7A0C
Mg in 721P
Mg in 6ZXH
Mg in 6ZXG