Magnesium »
PDB 8ec3-8elu »
8ehq »
Magnesium in PDB 8ehq: Mycobacterium Tuberculosis Paused Transcription Complex with Bacillus Subtilis Nusg
Enzymatic activity of Mycobacterium Tuberculosis Paused Transcription Complex with Bacillus Subtilis Nusg
All present enzymatic activity of Mycobacterium Tuberculosis Paused Transcription Complex with Bacillus Subtilis Nusg:
2.7.7.6;
2.7.7.6;
Other elements in 8ehq:
The structure of Mycobacterium Tuberculosis Paused Transcription Complex with Bacillus Subtilis Nusg also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Mycobacterium Tuberculosis Paused Transcription Complex with Bacillus Subtilis Nusg
(pdb code 8ehq). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Mycobacterium Tuberculosis Paused Transcription Complex with Bacillus Subtilis Nusg, PDB code: 8ehq:
In total only one binding site of Magnesium was determined in the Mycobacterium Tuberculosis Paused Transcription Complex with Bacillus Subtilis Nusg, PDB code: 8ehq:
Magnesium binding site 1 out of 1 in 8ehq
Go back to
Magnesium binding site 1 out
of 1 in the Mycobacterium Tuberculosis Paused Transcription Complex with Bacillus Subtilis Nusg
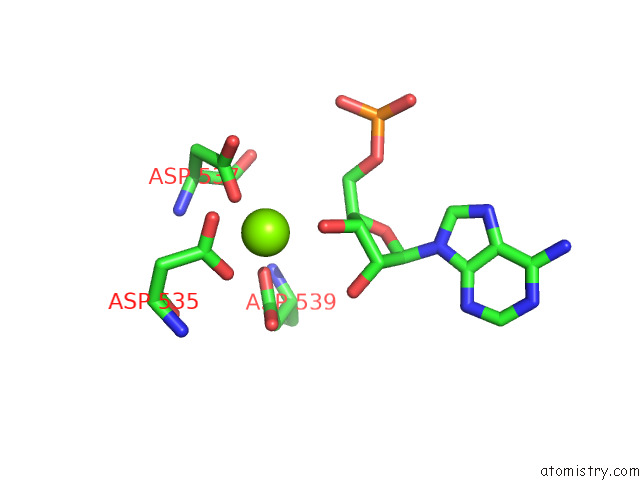
Mono view
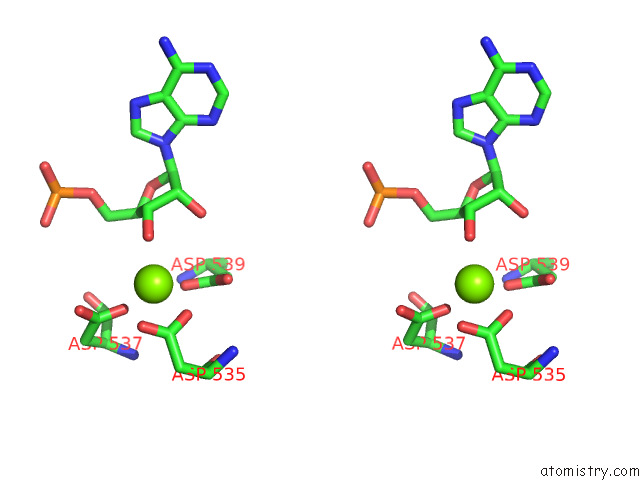
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Mycobacterium Tuberculosis Paused Transcription Complex with Bacillus Subtilis Nusg within 5.0Å range:
|
Reference:
R.K.Vishwakarma,
M.Z.Qayyum,
P.Babitzke,
K.S.Murakami.
Structural Basis of Nusg-Dependent Rna Polymerase Transcription Pausing in Bacteria To Be Published.
Page generated: Fri Aug 15 03:46:40 2025
Last articles
Mg in 8H53Mg in 8H68
Mg in 8H5Y
Mg in 8H4P
Mg in 8H5M
Mg in 8H4G
Mg in 8H4F
Mg in 8H4D
Mg in 8H4E
Mg in 8H4C