Magnesium »
PDB 8wp0-8x3y »
8x22 »
Magnesium in PDB 8x22: Hiv-1 Reverse Transcriptase Mutant Q151M/Y115F/F116Y/L74V:Dna:Dgtp Ternary Complex
Protein crystallography data
The structure of Hiv-1 Reverse Transcriptase Mutant Q151M/Y115F/F116Y/L74V:Dna:Dgtp Ternary Complex, PDB code: 8x22
was solved by
Y.Yasutake,
S.I.Hattori,
H.Mitsuya,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.81 / 2.31 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 285.227, 285.227, 96.075, 90, 90, 120 |
| R / Rfree (%) | 18.3 / 21.9 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Hiv-1 Reverse Transcriptase Mutant Q151M/Y115F/F116Y/L74V:Dna:Dgtp Ternary Complex
(pdb code 8x22). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 2 binding sites of Magnesium where determined in the Hiv-1 Reverse Transcriptase Mutant Q151M/Y115F/F116Y/L74V:Dna:Dgtp Ternary Complex, PDB code: 8x22:
Jump to Magnesium binding site number: 1; 2;
In total 2 binding sites of Magnesium where determined in the Hiv-1 Reverse Transcriptase Mutant Q151M/Y115F/F116Y/L74V:Dna:Dgtp Ternary Complex, PDB code: 8x22:
Jump to Magnesium binding site number: 1; 2;
Magnesium binding site 1 out of 2 in 8x22
Go back to
Magnesium binding site 1 out
of 2 in the Hiv-1 Reverse Transcriptase Mutant Q151M/Y115F/F116Y/L74V:Dna:Dgtp Ternary Complex
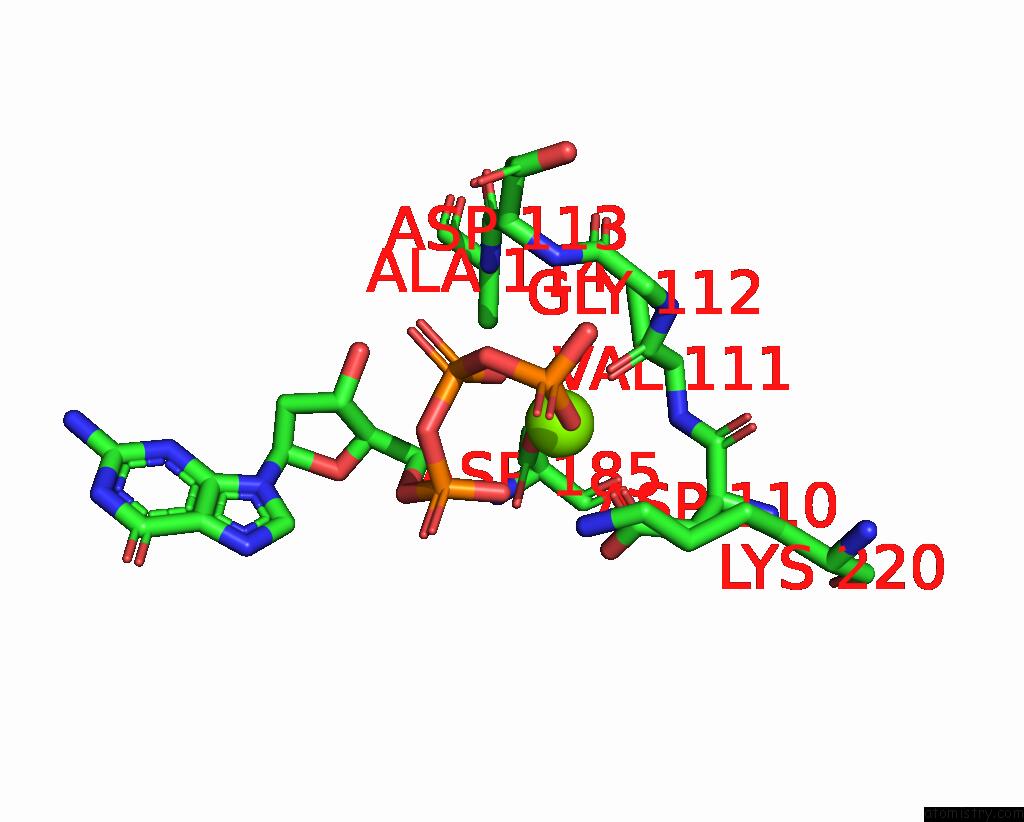
Mono view
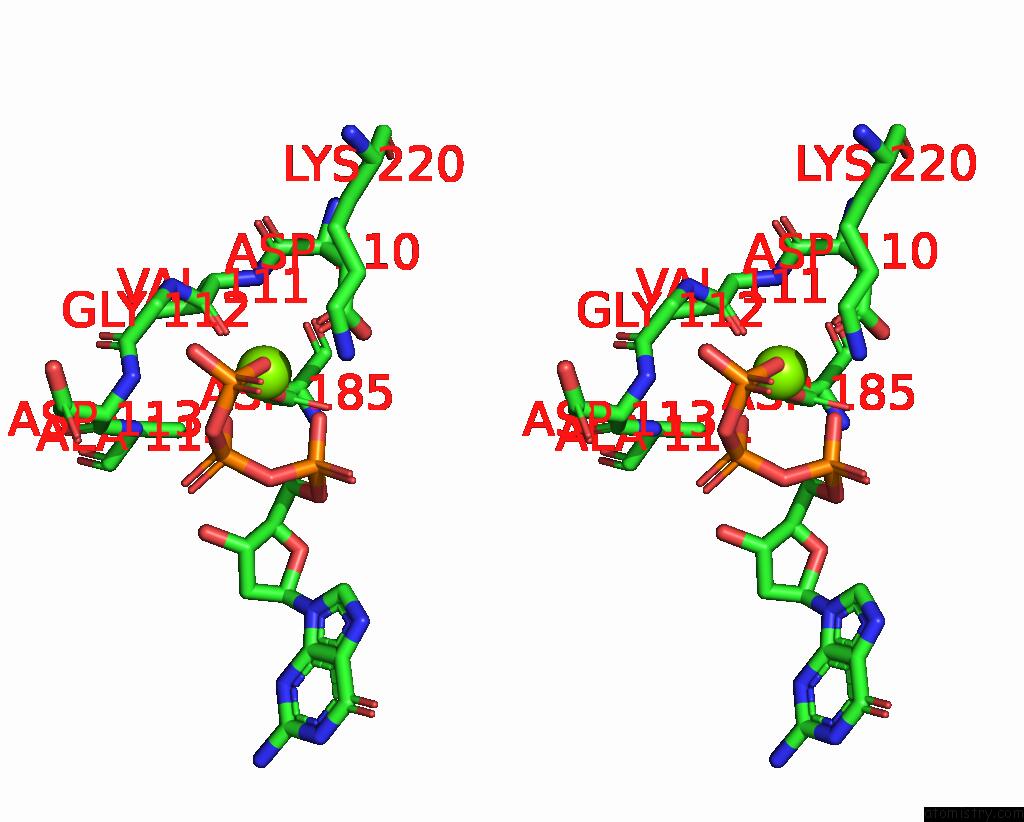
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Hiv-1 Reverse Transcriptase Mutant Q151M/Y115F/F116Y/L74V:Dna:Dgtp Ternary Complex within 5.0Å range:
|
Magnesium binding site 2 out of 2 in 8x22
Go back to
Magnesium binding site 2 out
of 2 in the Hiv-1 Reverse Transcriptase Mutant Q151M/Y115F/F116Y/L74V:Dna:Dgtp Ternary Complex
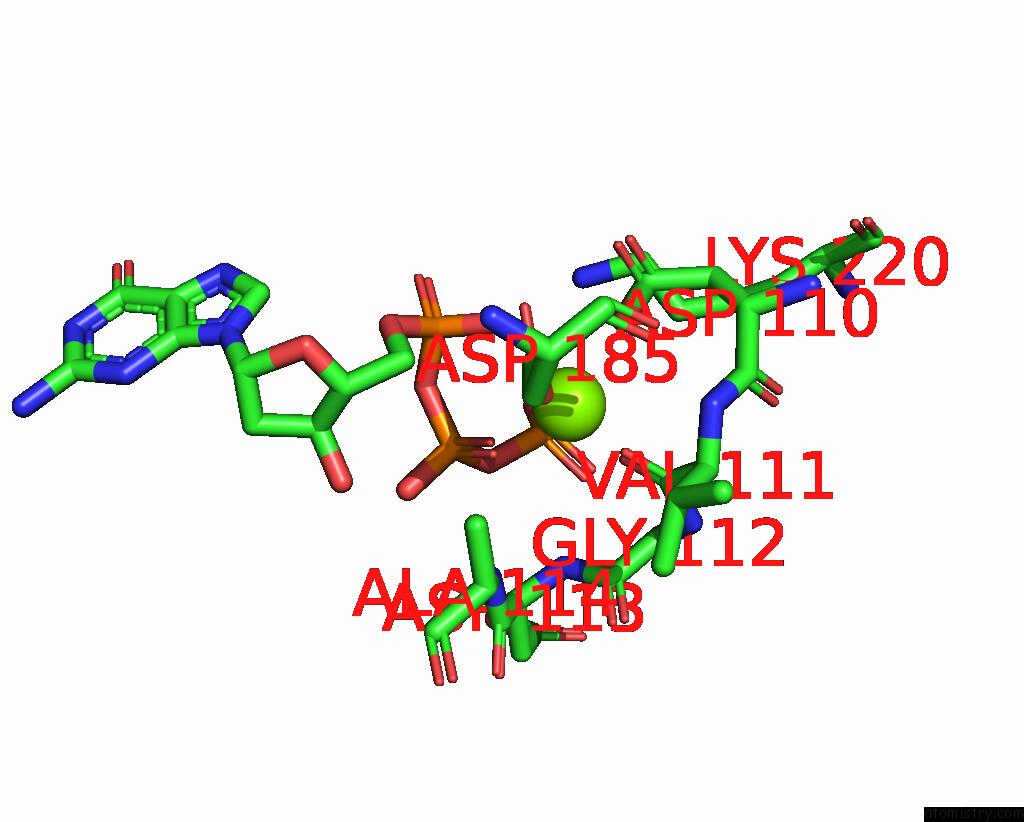
Mono view
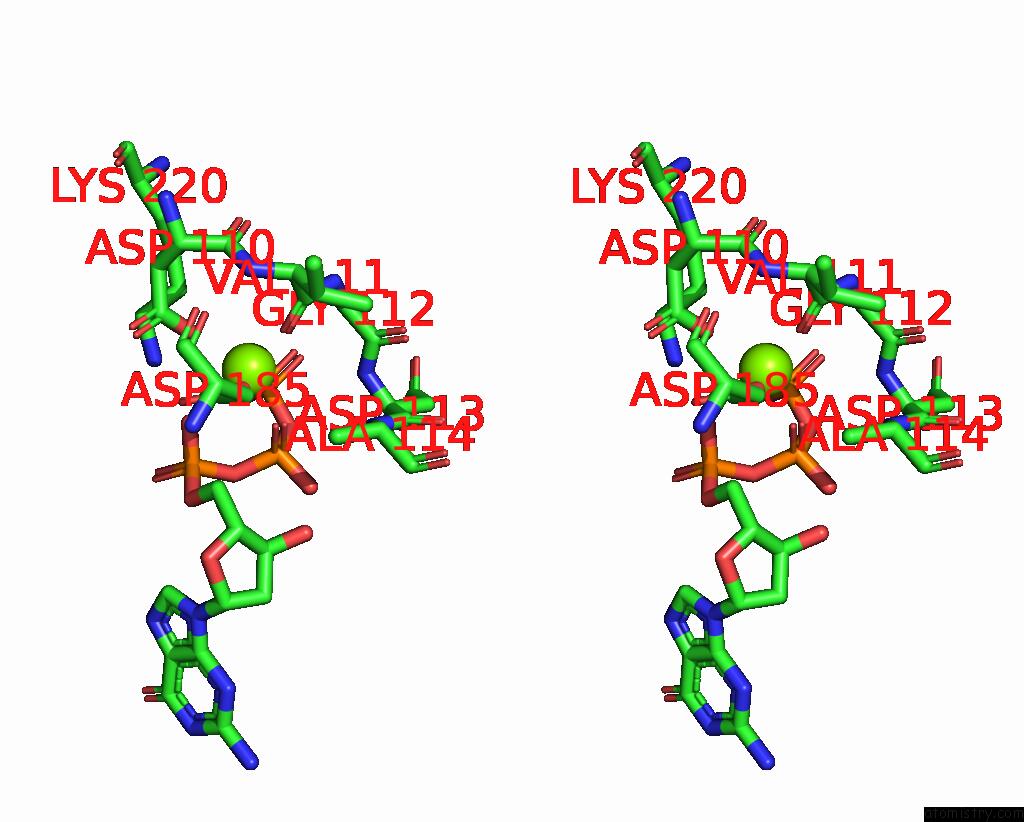
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Hiv-1 Reverse Transcriptase Mutant Q151M/Y115F/F116Y/L74V:Dna:Dgtp Ternary Complex within 5.0Å range:
|
Reference:
Y.Yasutake,
S.I.Hattori,
H.Kumamoto,
N.Tamura,
K.Maeda,
H.Mitsuya.
Deviated Binding of Anti-Hbv Nucleoside Analog E-Cfcp-Tp to the Reverse Transcriptase Active Site Attenuates the Effect of Drug-Resistant Mutations. Sci Rep V. 14 15742 2024.
ISSN: ESSN 2045-2322
PubMed: 38977798
DOI: 10.1038/S41598-024-66505-Z
Page generated: Fri Aug 15 20:07:21 2025
ISSN: ESSN 2045-2322
PubMed: 38977798
DOI: 10.1038/S41598-024-66505-Z
Last articles
Mg in 9GUPMg in 9GUR
Mg in 9GRE
Mg in 9GTK
Mg in 9GU5
Mg in 9GOB
Mg in 9GO5
Mg in 9GMZ
Mg in 9GQO
Mg in 9GMX