Magnesium »
PDB 2fx3-2g9y »
2g3d »
Magnesium in PDB 2g3d: Structure of S65G Y66A Gfp Variant After Spontaneous Peptide Hydrolysis
Protein crystallography data
The structure of Structure of S65G Y66A Gfp Variant After Spontaneous Peptide Hydrolysis, PDB code: 2g3d
was solved by
D.P.Barondeau,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 1.35 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.000, 62.600, 70.000, 90.00, 90.00, 90.00 |
| R / Rfree (%) | n/a / 21.5 |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Structure of S65G Y66A Gfp Variant After Spontaneous Peptide Hydrolysis
(pdb code 2g3d). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Structure of S65G Y66A Gfp Variant After Spontaneous Peptide Hydrolysis, PDB code: 2g3d:
In total only one binding site of Magnesium was determined in the Structure of S65G Y66A Gfp Variant After Spontaneous Peptide Hydrolysis, PDB code: 2g3d:
Magnesium binding site 1 out of 1 in 2g3d
Go back to
Magnesium binding site 1 out
of 1 in the Structure of S65G Y66A Gfp Variant After Spontaneous Peptide Hydrolysis
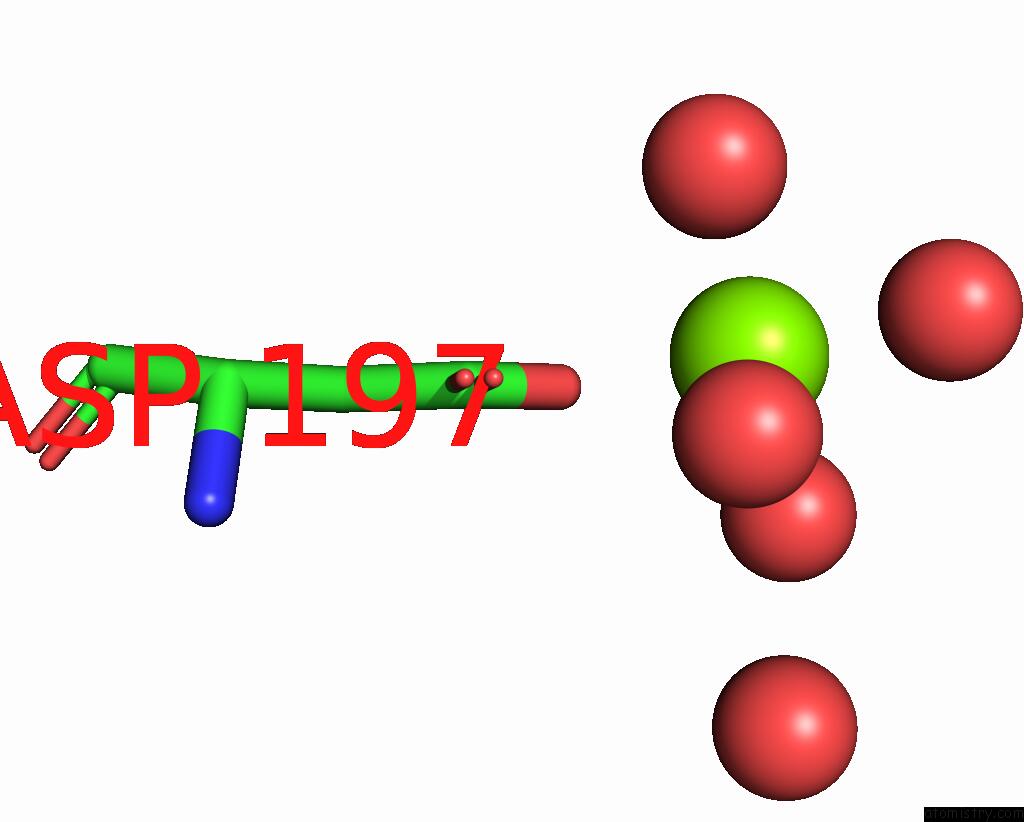
Mono view
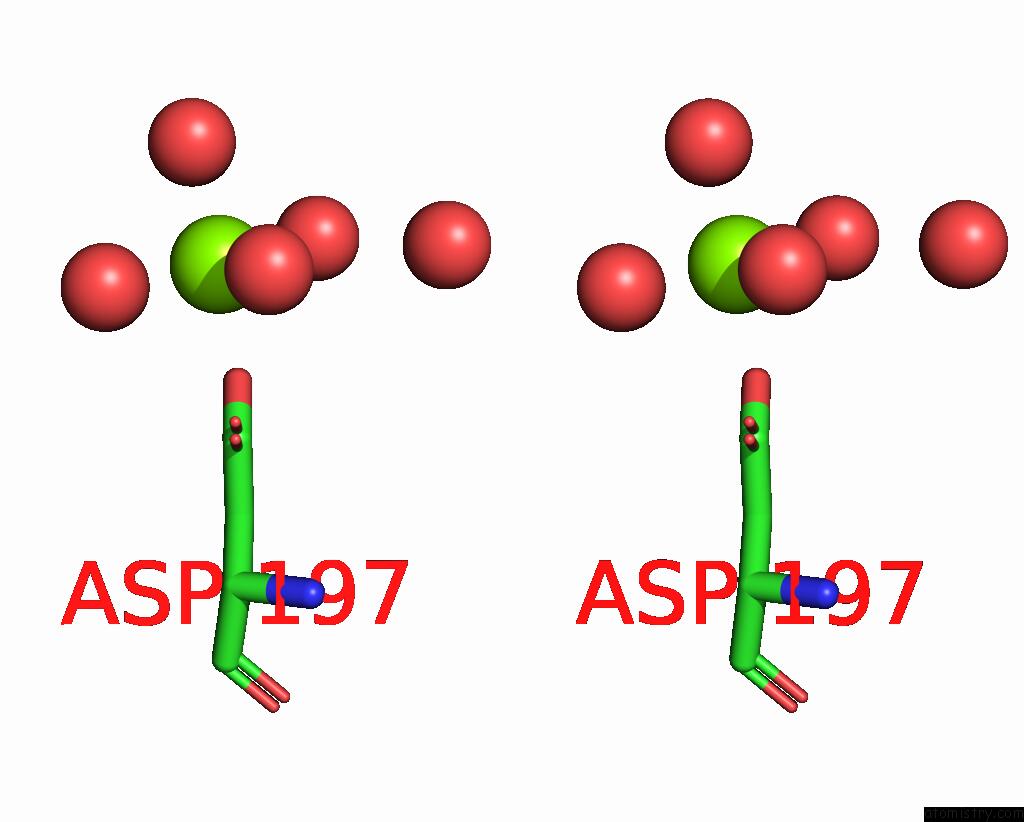
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Structure of S65G Y66A Gfp Variant After Spontaneous Peptide Hydrolysis within 5.0Å range:
|
Reference:
D.P.Barondeau,
C.J.Kassmann,
J.A.Tainer,
E.D.Getzoff.
Understanding Gfp Posttranslational Chemistry: Structures of Designed Variants That Achieve Backbone Fragmentation, Hydrolysis, and Decarboxylation. J.Am.Chem.Soc. V. 128 4685 2006.
ISSN: ISSN 0002-7863
PubMed: 16594705
DOI: 10.1021/JA056635L
Page generated: Tue Aug 13 23:22:21 2024
ISSN: ISSN 0002-7863
PubMed: 16594705
DOI: 10.1021/JA056635L
Last articles
Mg in 2A6EMg in 2A5Z
Mg in 2A5L
Mg in 2A5Y
Mg in 2A5J
Mg in 2A43
Mg in 2A5G
Mg in 2A5D
Mg in 2A5F
Mg in 2A42