Magnesium »
PDB 6nop-6o2r »
6nyy »
Magnesium in PDB 6nyy: Human M-Aaa Protease AFG3L2, Substrate-Bound
Other elements in 6nyy:
The structure of Human M-Aaa Protease AFG3L2, Substrate-Bound also contains other interesting chemical elements:
| Zinc | (Zn) | 6 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Human M-Aaa Protease AFG3L2, Substrate-Bound
(pdb code 6nyy). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total 3 binding sites of Magnesium where determined in the Human M-Aaa Protease AFG3L2, Substrate-Bound, PDB code: 6nyy:
Jump to Magnesium binding site number: 1; 2; 3;
In total 3 binding sites of Magnesium where determined in the Human M-Aaa Protease AFG3L2, Substrate-Bound, PDB code: 6nyy:
Jump to Magnesium binding site number: 1; 2; 3;
Magnesium binding site 1 out of 3 in 6nyy
Go back to
Magnesium binding site 1 out
of 3 in the Human M-Aaa Protease AFG3L2, Substrate-Bound
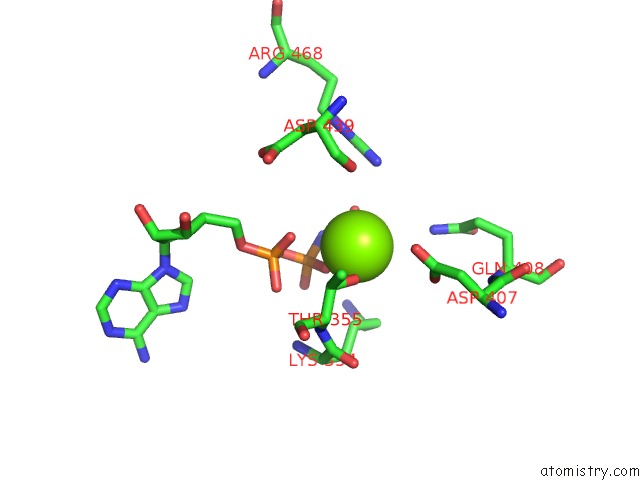
Mono view
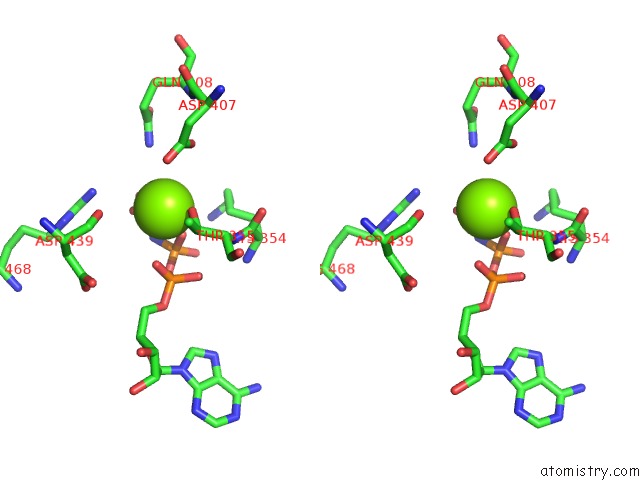
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Human M-Aaa Protease AFG3L2, Substrate-Bound within 5.0Å range:
|
Magnesium binding site 2 out of 3 in 6nyy
Go back to
Magnesium binding site 2 out
of 3 in the Human M-Aaa Protease AFG3L2, Substrate-Bound
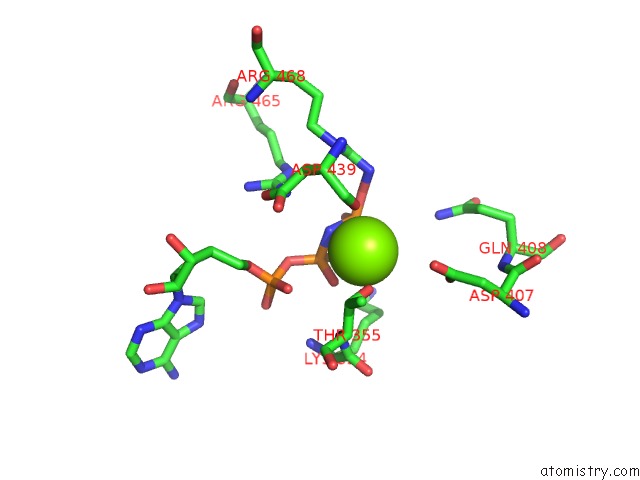
Mono view
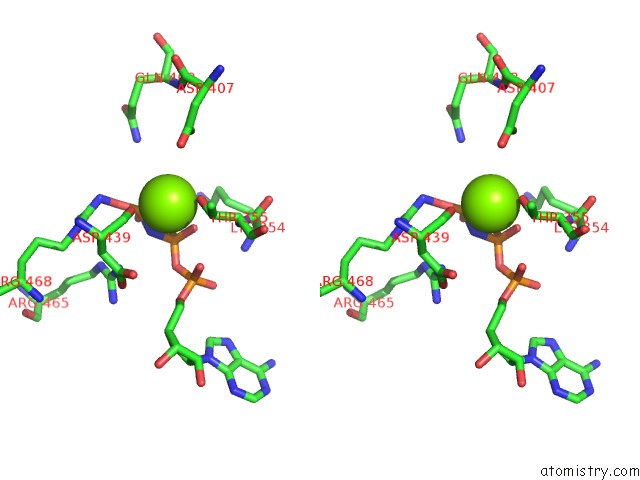
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 2 of Human M-Aaa Protease AFG3L2, Substrate-Bound within 5.0Å range:
|
Magnesium binding site 3 out of 3 in 6nyy
Go back to
Magnesium binding site 3 out
of 3 in the Human M-Aaa Protease AFG3L2, Substrate-Bound
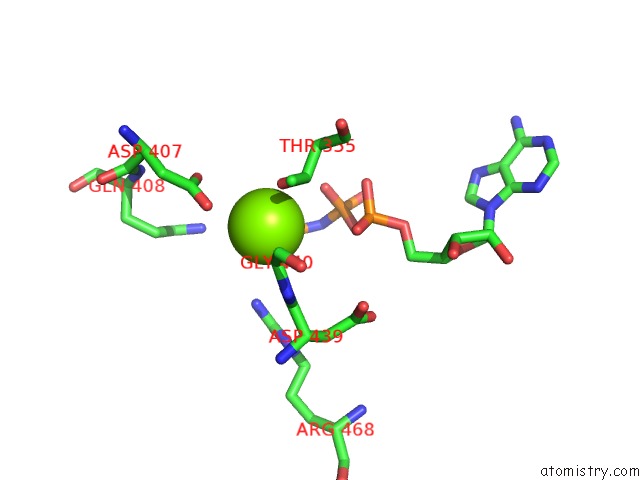
Mono view
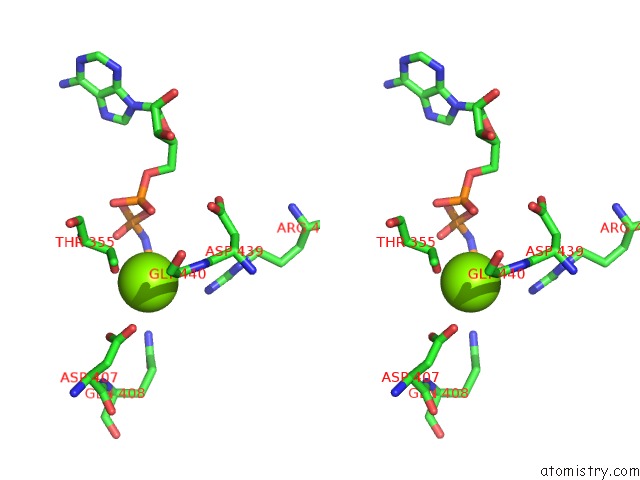
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 3 of Human M-Aaa Protease AFG3L2, Substrate-Bound within 5.0Å range:
|
Reference:
C.Puchades,
B.Ding,
A.Song,
R.L.Wiseman,
G.C.Lander,
S.E.Glynn.
Unique Structural Features of the Mitochondrial Aaa+ Protease AFG3L2 Reveal the Molecular Basis For Activity in Health and Disease. Mol.Cell V. 75 1073 2019.
ISSN: ISSN 1097-2765
PubMed: 31327635
DOI: 10.1016/J.MOLCEL.2019.06.016
Page generated: Tue Oct 1 12:56:18 2024
ISSN: ISSN 1097-2765
PubMed: 31327635
DOI: 10.1016/J.MOLCEL.2019.06.016
Last articles
Mg in 2CL6Mg in 2CL0
Mg in 2CK3
Mg in 2CL5
Mg in 2CJE
Mg in 2CJW
Mg in 2CJM
Mg in 2CJA
Mg in 2CJC
Mg in 2CIF