Magnesium »
PDB 8rav-8rjw »
8rea »
Magnesium in PDB 8rea: Cryo-Em Structure of Bacterial Rna Polymerase-SIGMA54 Initial Transcribing Complex - 5NT Post-Translocated Complex
Enzymatic activity of Cryo-Em Structure of Bacterial Rna Polymerase-SIGMA54 Initial Transcribing Complex - 5NT Post-Translocated Complex
All present enzymatic activity of Cryo-Em Structure of Bacterial Rna Polymerase-SIGMA54 Initial Transcribing Complex - 5NT Post-Translocated Complex:
2.7.7.6;
2.7.7.6;
Other elements in 8rea:
The structure of Cryo-Em Structure of Bacterial Rna Polymerase-SIGMA54 Initial Transcribing Complex - 5NT Post-Translocated Complex also contains other interesting chemical elements:
| Zinc | (Zn) | 2 atoms |
Magnesium Binding Sites:
The binding sites of Magnesium atom in the Cryo-Em Structure of Bacterial Rna Polymerase-SIGMA54 Initial Transcribing Complex - 5NT Post-Translocated Complex
(pdb code 8rea). This binding sites where shown within
5.0 Angstroms radius around Magnesium atom.
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of Bacterial Rna Polymerase-SIGMA54 Initial Transcribing Complex - 5NT Post-Translocated Complex, PDB code: 8rea:
In total only one binding site of Magnesium was determined in the Cryo-Em Structure of Bacterial Rna Polymerase-SIGMA54 Initial Transcribing Complex - 5NT Post-Translocated Complex, PDB code: 8rea:
Magnesium binding site 1 out of 1 in 8rea
Go back to
Magnesium binding site 1 out
of 1 in the Cryo-Em Structure of Bacterial Rna Polymerase-SIGMA54 Initial Transcribing Complex - 5NT Post-Translocated Complex
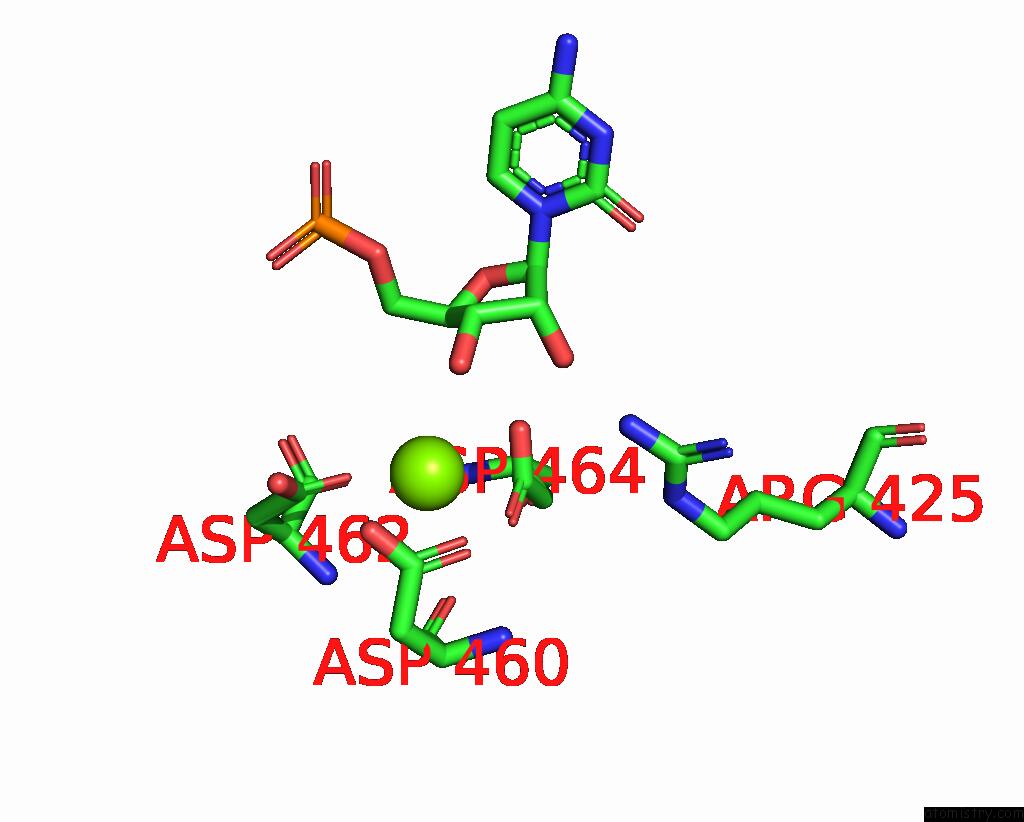
Mono view
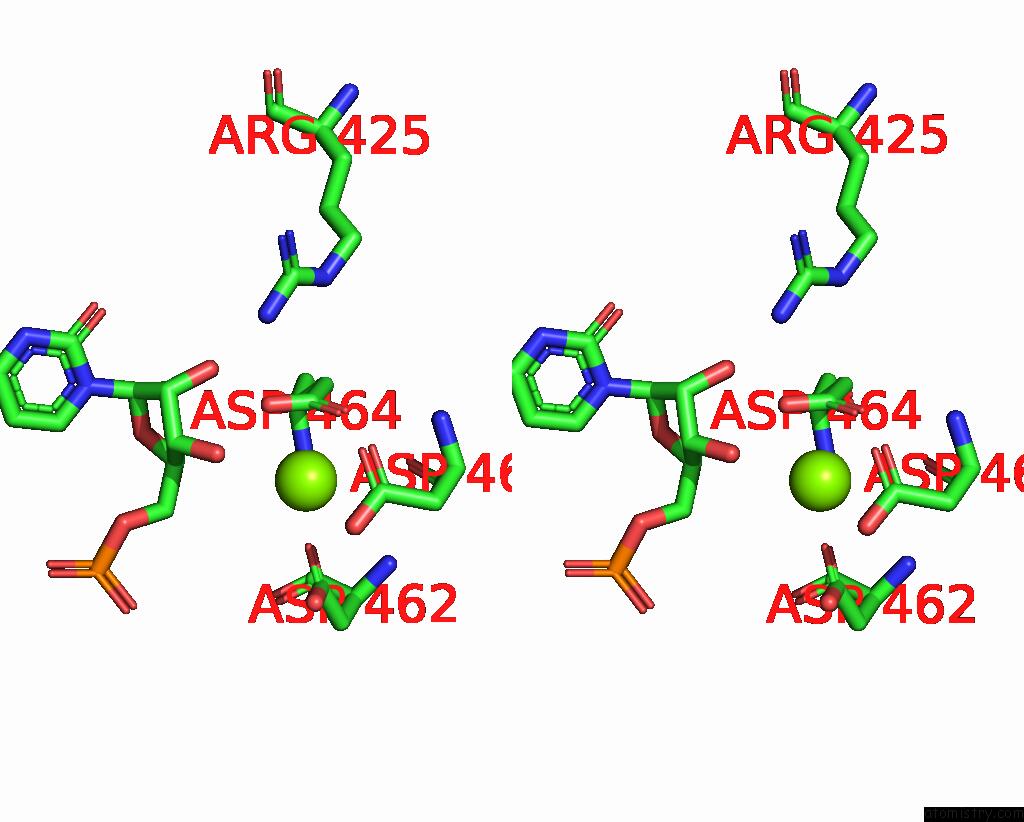
Stereo pair view

Mono view

Stereo pair view
A full contact list of Magnesium with other atoms in the Mg binding
site number 1 of Cryo-Em Structure of Bacterial Rna Polymerase-SIGMA54 Initial Transcribing Complex - 5NT Post-Translocated Complex within 5.0Å range:
|
Reference:
F.Gao,
F.Ye,
B.Zhang,
N.Cronin,
M.Buck,
X.Zhang.
Structural Basis of Sigma 54 Displacement and Promoter Escape in Bacterial Transcription. Proc.Natl.Acad.Sci.Usa V. 121 70120 2024.
ISSN: ESSN 1091-6490
PubMed: 38170755
DOI: 10.1073/PNAS.2309670120
Page generated: Fri Aug 15 13:57:42 2025
ISSN: ESSN 1091-6490
PubMed: 38170755
DOI: 10.1073/PNAS.2309670120
Last articles
Mg in 9J0OMg in 9IZ1
Mg in 9J0N
Mg in 9IZ4
Mg in 9IZQ
Mg in 9IZP
Mg in 9IZM
Mg in 9IYP
Mg in 9IY0
Mg in 9IZ2